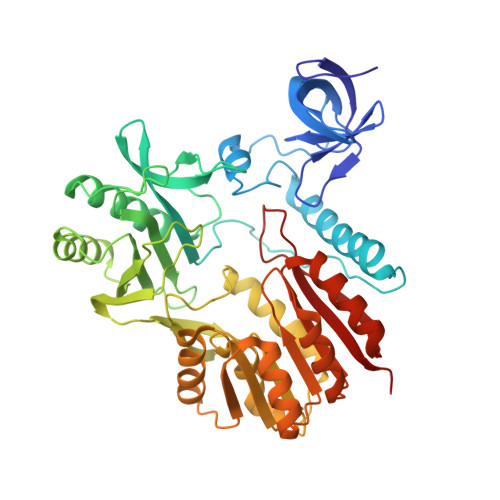
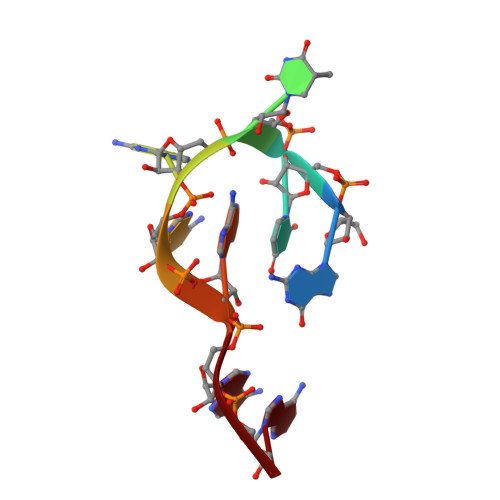
Unveiling the structural features that determine the dual methyltransferase activities of Streptococcus pneumoniae RlmCD
Jiang, Y., Yu, H., Li, F., Cheng, L., Zhu, L., Shi, Y., Gong, Q.(2018) PLoS Pathog 14: e1007379-e1007379
- PubMed: 30388185
- DOI: https://doi.org/10.1371/journal.ppat.1007379
- Primary Citation of Related Structures:
5ZQ0, 5ZQ1, 5ZQ8, 5ZTH - PubMed Abstract:
Methyltransferase RlmCD was previously shown to be responsible for the introduction of C5 methylation at both U747 and U1939 of the 23S ribosomal RNA in Streptococcus pneumoniae. Intriguingly, its structural homologue, RumA, can only catalyze the methylation of U1939, while RlmC is the dedicated enzyme for m5U747 in Escherichia coli. In this study, we describe the structure of RlmCD in complex with its cofactor and the RNA substrate containing U747 at 2.00 Å or U1939 at 3.10 Å. We demonstrate that multiple structural features collaborate to establish the dual enzymatic activities of RlmCD. Of them, the side-chain rearrangement of F145 was observed to be an unusual mechanism through which RlmCD can discriminate between U747- and U1939-containing RNA substrate by switching the intermolecular aromatic stacking between protein and RNA on/off. An in-vitro methyltransferase assay and electrophoretic mobility shift assay were performed to validate these findings. Overall, our complex structures allow for a better understanding of the dual-functional mechanism of RlmCD, suggesting useful implications for the evolution of the RumA-type enzyme and the potential development of antibiotic drugs against S. pneumoniae.
Organizational Affiliation:
Hefei National Laboratory for Physical Science at the Microscale, School of Life Sciences, University of Science and Technology of China, Hefei, Anhui, China.