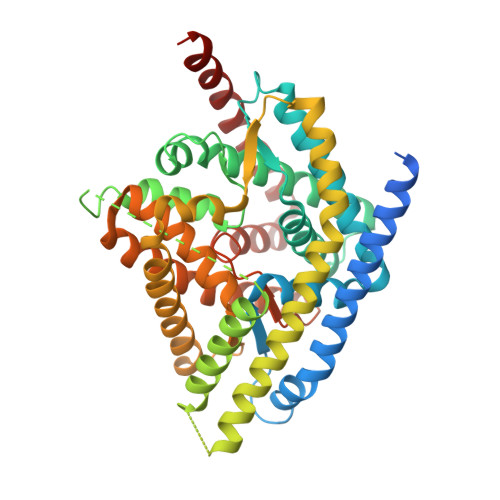
Inward-facing conformation of l-ascorbate transporter suggests an elevator mechanism
Luo, P., Dai, S., Zeng, J., Duan, J., Shi, H., Wang, J.(2018) Cell Discov 4: 35-35
- PubMed: 30038796
- DOI: https://doi.org/10.1038/s41421-018-0037-y
- Primary Citation of Related Structures:
5ZOV - PubMed Abstract:
Various bacteria can ferment vitamin C (l-ascorbate) under anaerobic conditions via the phosphoenolpyruvate-dependent phosphotransferase system (PTS). The PTS asc system is composed of two soluble energy-coupling proteins (EI and HPr) and an enzyme II complex (EIIA, EIIB, and EIIC) for the anaerobic uptake of ascorbate and its phosphorylation to l-ascorbate 6-phosphate in vivo. Crystal structures of the ascorbate-bound EIIC component from Escherichia coli are available in outward-open and occluded conformations, suggesting a possible elevator mechanism of membrane transport. Despite these advances, it remains unclear how EIIC actually transports the substrate across the membrane and interacts with EIIB, which transfers its phosphate group to the EIIC-embedding ascorbate. Here, we present the crystal structure of the EIIC asc component from Pasteurella multocida in the inward-facing conformation. By comparing three conformational states, we confirmed the original proposed model: the ascorbate translocation can be achieved by a rigid-body movement of the substrate-binding core domain relative to the V motif domain, which brings along the transmembrane helices TM2 and TM7 of the V motif domain to undergo a winding at the pivotal positions. Together with an in vivo transport assay, we completed the picture of the transport cycle of the ascorbate superfamily of membrane-spanning EIIC components of the PTS system.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, 100084 Beijing, China.