Structural basis of protein-protein interactions between a trans-acting acyltransferase and acyl carrier protein in polyketide disorazole biosynthesis
Miyanaga, A., Ouchi, R., Ishikawa, F., Goto, E., Tanabe, G., Kudo, F., Eguchi, T.(2018) J Am Chem Soc 140: 7970-7978
- PubMed: 29870659
- DOI: https://doi.org/10.1021/jacs.8b04162
- Primary Citation of Related Structures:
5ZK4 - PubMed Abstract:
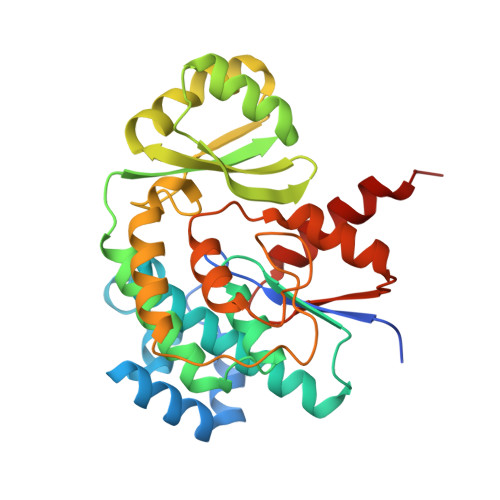
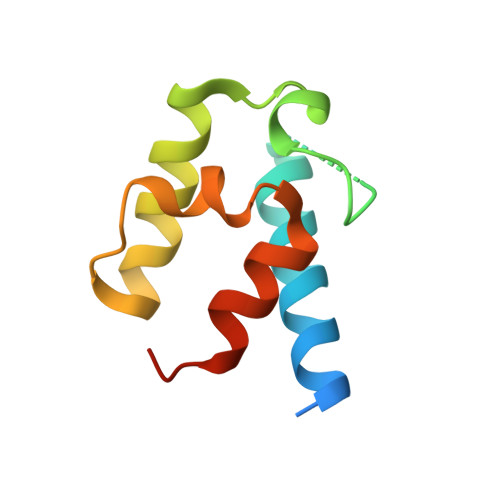
Acyltransferases (ATs) are responsible for the selection and incorporation of acyl building blocks in the biosynthesis of various polyketide natural products. The trans-AT modular polyketide synthases have a discrete trans-acting AT for the loading of an acyl unit onto the acyl carrier protein (ACP) located within each module. Despite the importance of protein-protein interactions between ATs and ACPs in trans-AT assembly lines, the dynamic actions of ACPs and trans-acting ATs remain largely uncharacterized because of the inherently transient nature of ACP-enzyme interactions. Herein, we report the crystal structure of the AT-ACP complex of disorazole trans-AT polyketide synthase. We used a bromoacetamide pantetheine cross-linking probe in combination with a Cys mutation to trap the transient AT-ACP complex, allowing the determination of the crystal structure of the disorazole AT-ACP complex at 2.03 Å resolution. On the basis of the cross-linked AT-ACP complex structure, ACP residues recognized by trans-acting AT were identified and validated by mutational studies, which demonstrated that the disorazole AT recognizes the loop 1 and helix III' residues of disorazole ACP. The disorazole AT-ACP complex structure presents a foundation for defining the dynamic processes associated with trans-acting ATs and provides detailed mechanistic insights into their ability to recognize ACPs.
Organizational Affiliation:
Department of Chemistry , Tokyo Institute of Technology , 2-12-1, O-okayama , Meguro-ku, Tokyo 152-8551 , Japan.