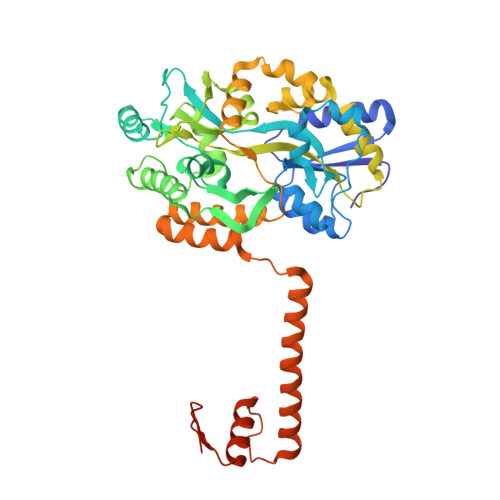
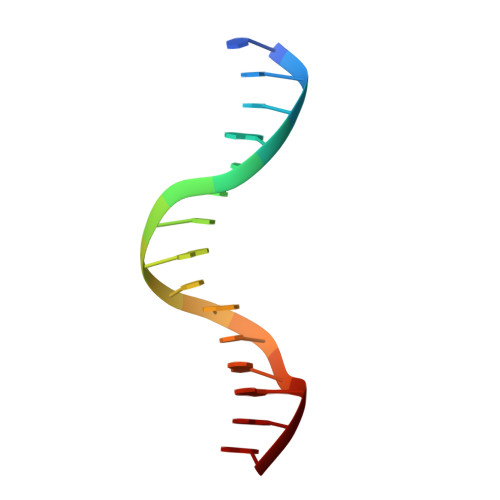
Structural basis for brassinosteroid response by BIL1/BZR1.
Nosaki, S., Miyakawa, T., Xu, Y., Nakamura, A., Hirabayashi, K., Asami, T., Nakano, T., Tanokura, M.(2018) Nat Plants 4: 771-776
- PubMed: 30287951
- DOI: https://doi.org/10.1038/s41477-018-0255-1
- Primary Citation of Related Structures:
5ZD4 - PubMed Abstract:
BRZ-INSENSITIVE-LONG HYPOCOTYL 1 (BIL1)/BRASSINAZOLE-RESISTANT 1 (BZR1) is a master transcription factor of brassinosteroid (BR) signalling. The varieties of nucleobase recognition of the NN-BRRE-core motif (NNCGTG), one of variant G-box motifs, distinguish BIL1/BZR1 from basic helix-loop-helix transcription factors, underlying the specific regulation of BR-responsive genes. Here, we show the non-canonical bHLH dimer formation of BIL1/BZR1 to optimize the interaction network with DNA and the orientation of a key residue for NN-BRRE-core motif recognition.
Organizational Affiliation:
Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan.