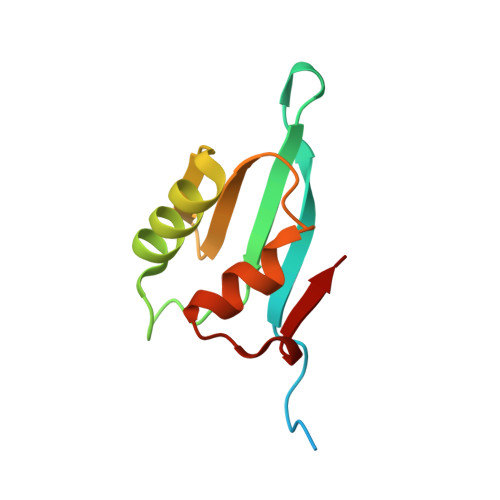
Archaeal and eukaryal translation initiation factor 1 differ in their RNA interacting loops.
Gogoi, P., Kanaujia, S.P.(2018) FEBS Lett 592: 1602-1610
- PubMed: 29608219
- DOI: https://doi.org/10.1002/1873-3468.13044
- Primary Citation of Related Structures:
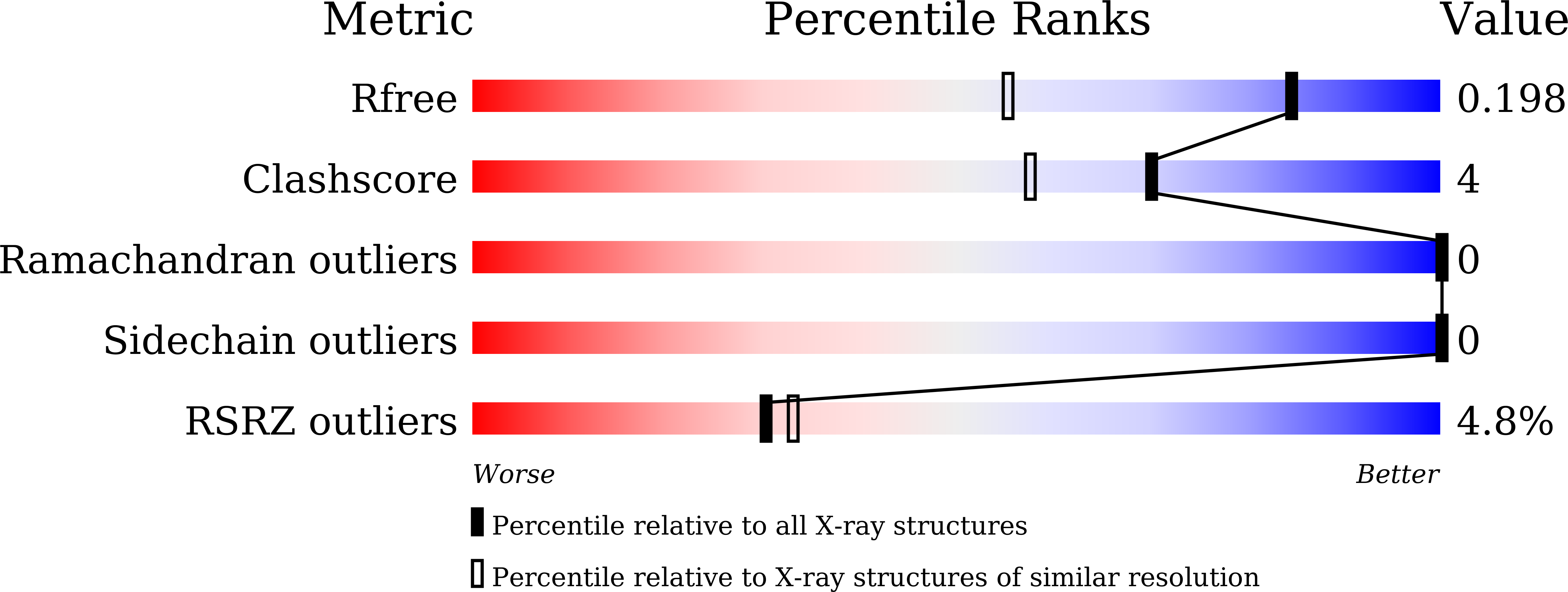
5ZCY - PubMed Abstract:
The archaeal translation initiation factor 1 (aIF1) is reported to be functionally homologous to the eukaryotic translation initiation factor 1 (eIF1). However, lack of a structural comparison between aIF1 and eIF1 has limited our understanding of the structural (dis)similarities. Herein, we have determined the three-dimensional crystal structure of an open reading frame PH1771.1 encoding aIF1 in Pyrococcus horikoshii OT3. Results reveal that although aIF1 has low sequence similarity with eIF1, high structural homology exists between the two proteins. Nonetheless, notable critical differences between aIF1 and eIF1 could still be perceived at the β 1 -β 2 basic loop, the acidic loop and the solvent-exposed surface. These differences might lead to a slightly divergent mode of action of aIF1 during archaeal translation initiation.
Organizational Affiliation:
Department of Biosciences and Bioengineering, Indian Institute of Technology Guwahati, Assam, India.