Structure of a [NiFe] hydrogenase maturation protease HycI provides insights into its substrate selectivity
Kwon, S., Nishitani, Y., Hirao, Y., Kanai, T., Atomi, H., Miki, K.(2018) Biochem Biophys Res Commun 498: 782-788
- PubMed: 29526754
- DOI: https://doi.org/10.1016/j.bbrc.2018.03.058
- Primary Citation of Related Structures:
5ZBY - PubMed Abstract:
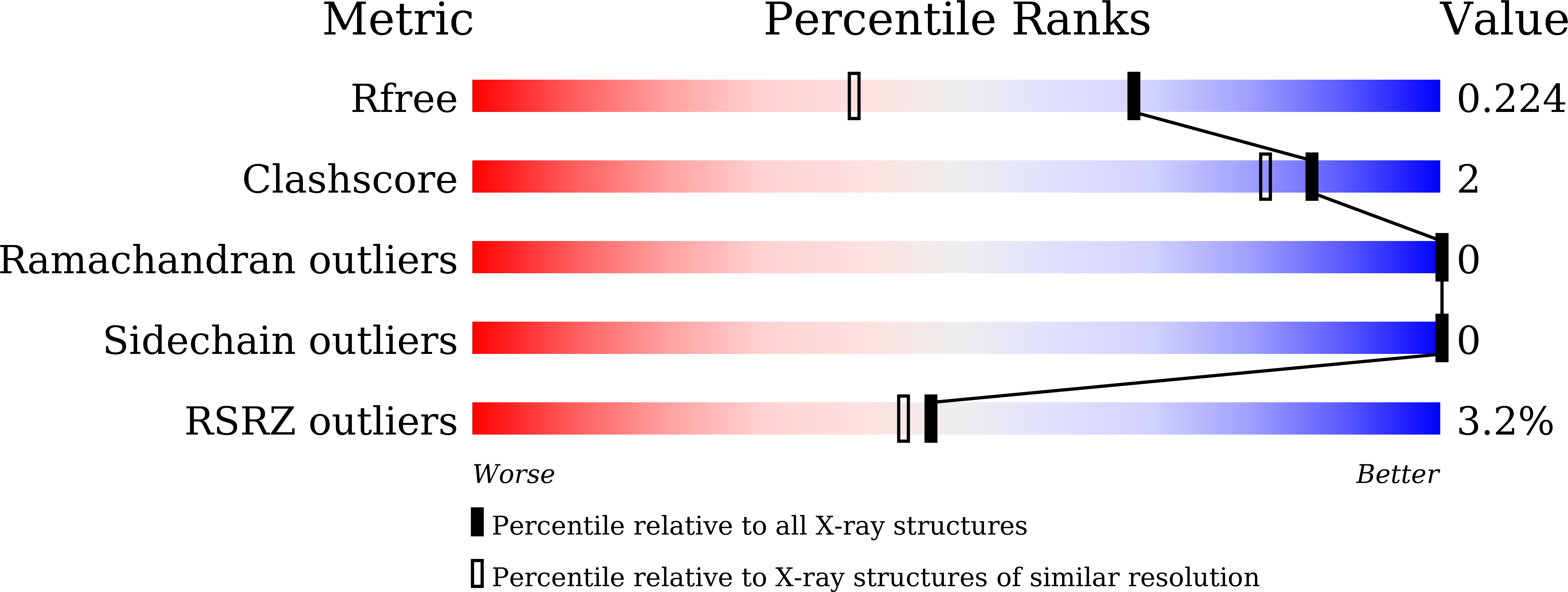
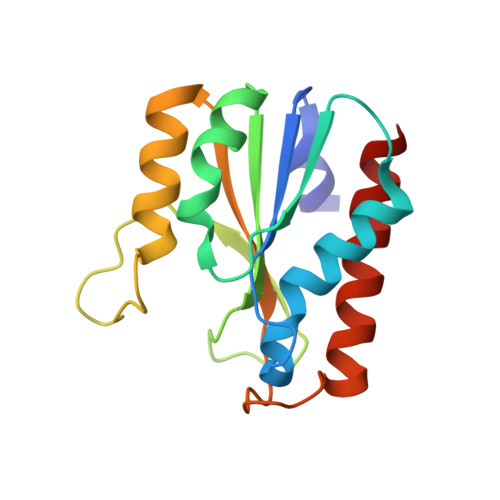
The immature large subunit of [NiFe] hydrogenases undergoes C-terminal cleavage by a specific protease in the final step of the post-translational process before assembly with other subunits. It has been reported that the [NiFe] hydrogenase maturation protease HycI from Thermococcus kodakarensis (TkHycI) has the catalytic ability to target the membrane-bound hydrogenase large subunit MbhL from T. kodakarensis. However, the detailed mechanism of its substrate recognition remains elusive. We determined the crystal structure of TkHycI at 1.59 Å resolution to clarify how TkHycI recognizes its own substrate MbhL. Although the overall structure of TkHycI is similar to that of its homologous protease TkHybD, TkHycI adopts a larger loop than TkHybD, thereby creating a broad and deep cleft. We analyzed the structural properties of the TkHycI cleft probably involved in its substrate recognition. Our findings provide novel and profound insights into the substrate selectivity of TkHycI.
Organizational Affiliation:
Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto, 606-8502, Japan.