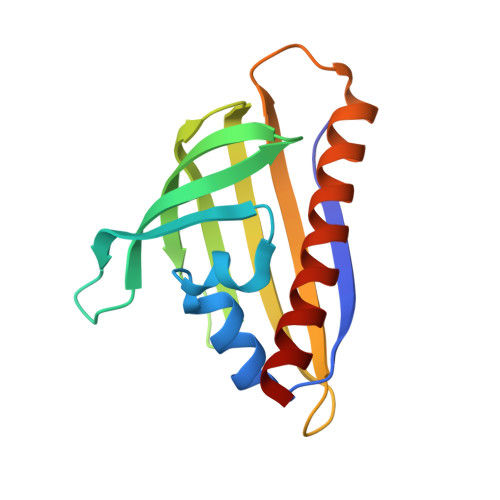
Structural and genetic analysis of START superfamily protein MSMEG_0129 from Mycobacterium smegmatis.
Zheng, S., Zhou, Y., Fleming, J., Zhou, Y., Zhang, M., Li, S., Li, H., Sun, B., Liu, W., Bi, L.(2018) FEBS Lett 592: 1445-1457
- PubMed: 29512898
- DOI: https://doi.org/10.1002/1873-3468.13024
- Primary Citation of Related Structures:
5Z8O - PubMed Abstract:
Mycobacterium tuberculosis is a notorious pathogen that continues to threaten human health. Rv0164, an antigen of both T- and B cells conserved across mycobacteria, and MSMEG_0129, its close homolog in Mycobacterium smegmatis, are predicted members of the START domain superfamily, but their molecular function is unknown. Here, gene knockout studies demonstrate MSMEG_0129 is essential for bacterial growth, suggesting Rv0164 may be a potential drug target. The MSMEG_0129 crystal structure determined at 1.95 Å reveals a fold similar to that in polyketide aromatase/cyclases ZhuI and TcmN from Streptomyces sp. Structural comparisons and docking simulations, however, infer that MSMEG_0129 and Rv0164 are unlikely to catalyze polyketide aromatization/cyclization, but probably play an irreplaceable role during mycobacterial growth, for example, in lipid transfer during cell envelope synthesis.
Organizational Affiliation:
School of Stomatology and Medicine, Foshan University, China.