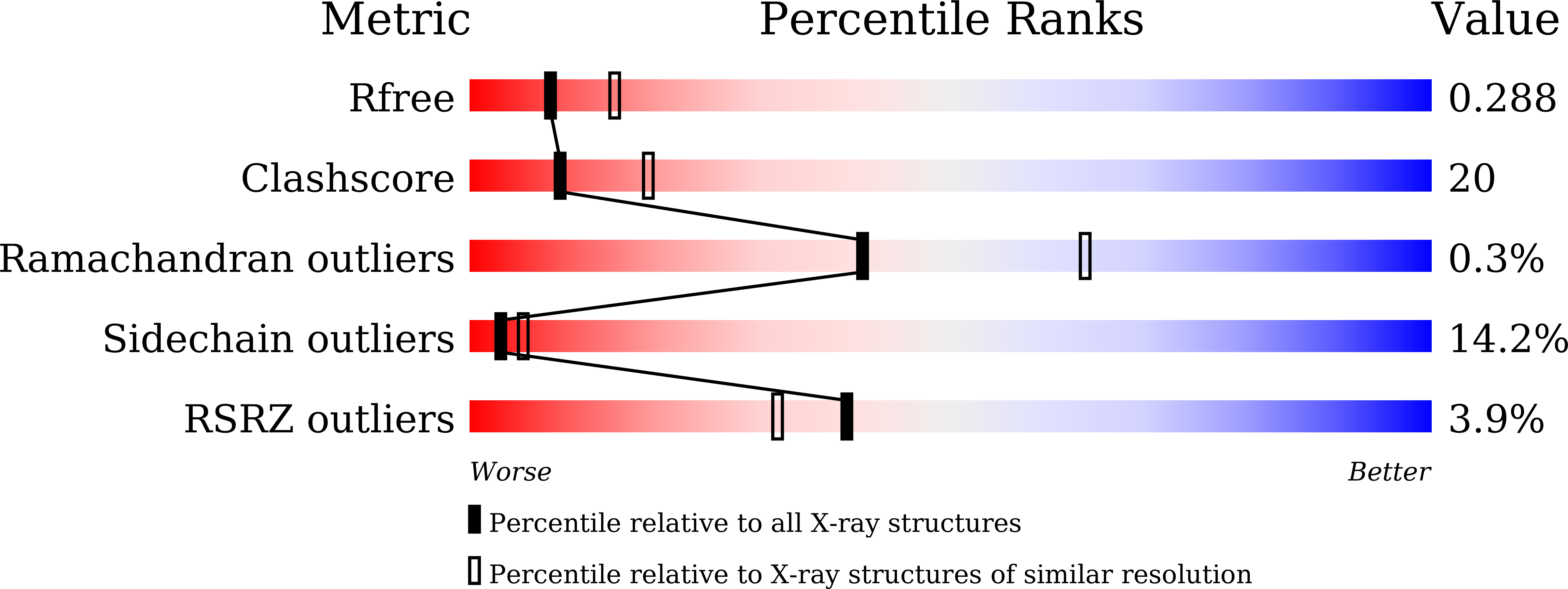
Vibrio cholerae LMWPTP-2 display unique surface charge and grooves around the active site: Indicative of distinctive substrate specificity and scope to design specific inhibitor.
Chatterjee, S., Nath, S., Ghosh, B., Sen, U.(2019) Biochim Biophys Acta Proteins Proteom 1867: 114-124
- PubMed: 30447286
- DOI: https://doi.org/10.1016/j.bbapap.2018.11.003
- Primary Citation of Related Structures:
5Z3M - PubMed Abstract:
Low molecular weight protein tyrosine phosphatases (LMWPTPs) are ubiquitously found as small cytoplasmic enzymes which act on phospho-tyrosine containing proteins that are engaged in various cellular functions. Vibrio cholerae O395 contains two LMWPTPs having widely different sequence. Phylogenetic analysis based on a non redundant set of 124 LMWPTP sequences, designate that LMWPTP-2 from Vibrio choleraeO395 (VcLMWPTP-2) is a single taxon. We have determined the crystal structure of VcLMWPTP-2 at 2.6 Å with MOPS bound in the active site. Tertiary structure analysis indicates that VcLMWPTP-2 forms dimer. Studies in solution state also confirm exclusive presence of a dimeric form. Kinetic studies demonstrate that VcLMWPTP-2 dimer is catalytically active while inactivation through oligomerisation was reported as one of the regulatory mechanism in case of mammalian LMWPTP viz., Bos taurus LMWPTP, BPTP. Kinetic studies using p-nitrophenyl phosphate (p-NPP) as a substrate demonstrate active participation of both the P-loop cysteine in catalysis. Vicinal Cys17, in addition plays a role of protecting the catalytic Cys12 under oxidative stress. Structural analysis and MD simulations allowed us to propose the role of several conserved residues around the active site. Distribution of surface charges and grooves around the active site delineates unique features of VcLMWPTP-2 which could be utilized to design specific inhibitor.
Organizational Affiliation:
Crystallography and Molecular Biology Division, Saha Institute of Nuclear Physics, HBNI, 1/AF Bidhan Nagar, Kolkata 700064, India.