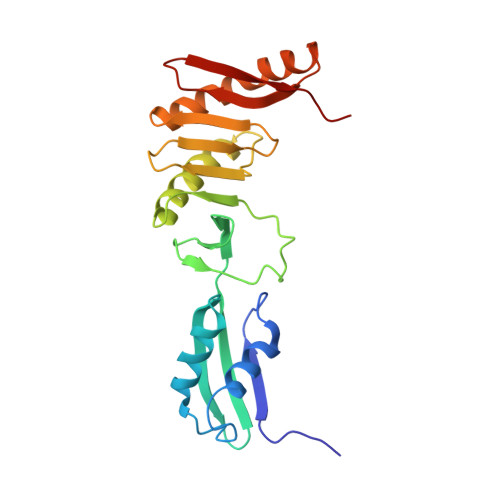
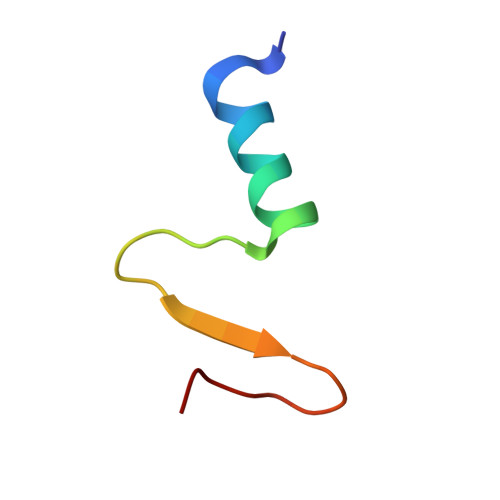
Structural Insights into the FtsQ/FtsB/FtsL Complex, a Key Component of the Divisome.
Choi, Y., Kim, J., Yoon, H.J., Jin, K.S., Ryu, S., Lee, H.H.(2018) Sci Rep 8: 18061-18061
- PubMed: 30584256
- DOI: https://doi.org/10.1038/s41598-018-36001-2
- Primary Citation of Related Structures:
5Z2W - PubMed Abstract:
Bacterial cell division is a fundamental process that results in the physical separation of a mother cell into two daughter cells and involves a set of proteins known as the divisome. Among them, the FtsQ/FtsB/FtsL complex was known as a scaffold protein complex, but its overall structure and exact function is not precisely known. In this study, we have determined the crystal structure of the periplasmic domain of FtsQ in complex with the C-terminal fragment of FtsB, and showed that the C-terminal region of FtsB is a key binding region of FtsQ via mutational analysis in vitro and in vivo. We also obtained the solution structure of the periplasmic FtsQ/FtsB/FtsL complex by small angle X-ray scattering (SAXS), which reveals its structural organization. Interestingly, the SAXS and analytical gel filtration data showed that the FtsQ/FtsB/FtsL complex forms a 2:2:2 heterohexameric assembly in solution with the "Y" shape. Based on the model, the N-terminal directions of FtsQ and the FtsB/FtsL complex should be opposite, suggesting that the Y-shaped FtsQ/FtsB/FtsL complex might fit well into the curved membrane for membrane anchoring.
Organizational Affiliation:
Department of Chemistry, College of Natural Sciences, Seoul National University, Seoul, 08826, Korea.