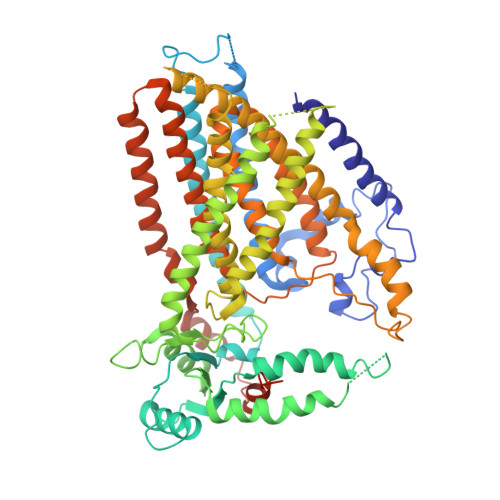
Structure of the mechanosensitive OSCA channels.
Zhang, M., Wang, D., Kang, Y., Wu, J.X., Yao, F., Pan, C., Yan, Z., Song, C., Chen, L.(2018) Nat Struct Mol Biol 25: 850-858
- PubMed: 30190597
- DOI: https://doi.org/10.1038/s41594-018-0117-6
- Primary Citation of Related Structures:
5Z1F, 6JPF - PubMed Abstract:
Mechanosensitive ion channels convert mechanical stimuli into a flow of ions. These channels are widely distributed from bacteria to higher plants and humans, and are involved in many crucial physiological processes. Here we show that two members of the OSCA protein family in Arabidopsis thaliana, namely AtOSCA1.1 and AtOSCA3.1, belong to a new class of mechanosensitive ion channels. We solve the structure of the AtOSCA1.1 channel at 3.5-Å resolution and AtOSCA3.1 at 4.8-Å resolution by cryo-electron microscopy. OSCA channels are symmetric dimers that are mediated by cytosolic inter-subunit interactions. Strikingly, they have structural similarity to the mammalian TMEM16 family proteins. Our structural analysis accompanied with electrophysiological studies identifies the ion permeation pathway within each subunit and suggests a conformational change model for activation.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, Institute of Molecular Medicine, Peking University, Beijing Key Laboratory of Cardiometabolic Molecular Medicine, Beijing, China.