Structural analysis of fungal CENP-H/I/K homologs reveals a conserved assembly mechanism underlying proper chromosome alignment.
Hu, L., Huang, H., Hei, M., Yang, Y., Li, S., Liu, Y., Dou, Z., Wu, M., Li, J., Wang, G.Z., Yao, X., Liu, H., He, X., Tian, W.(2019) Nucleic Acids Res 47: 468-479
- PubMed: 30407575
- DOI: https://doi.org/10.1093/nar/gky1108
- Primary Citation of Related Structures:
5Z07, 5Z08 - PubMed Abstract:
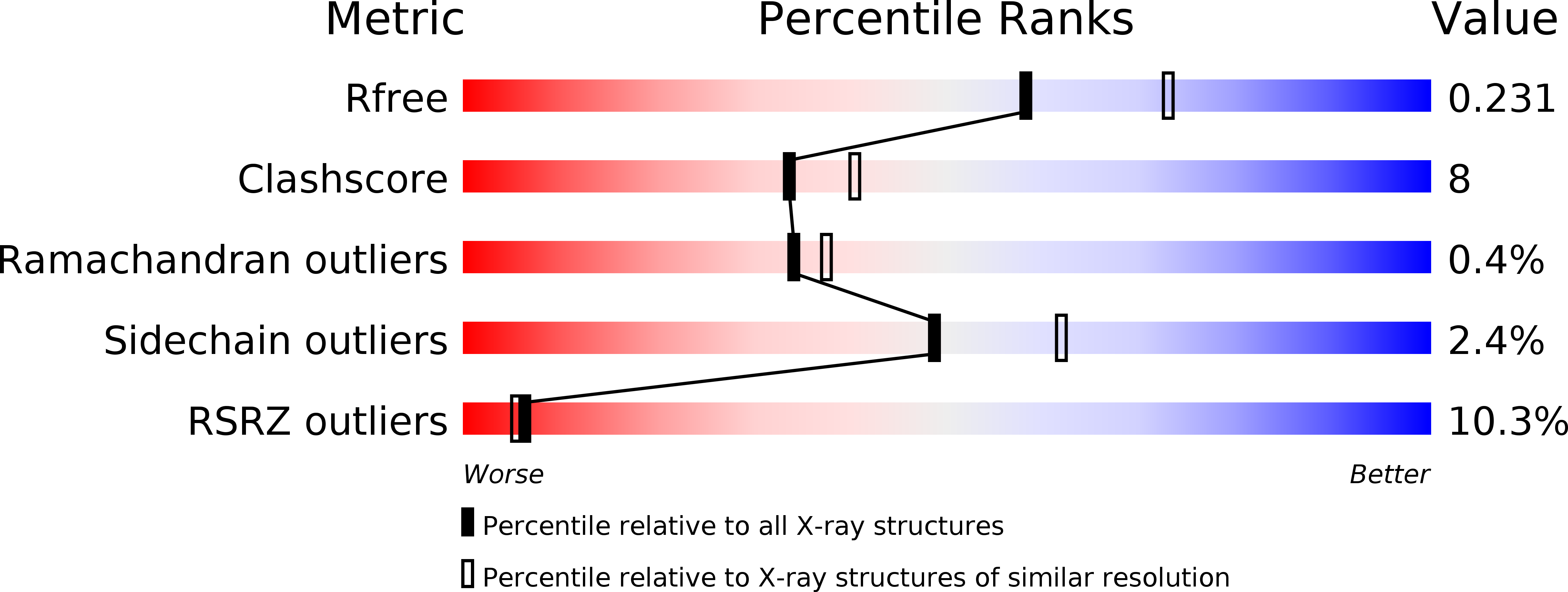
The kinetochore is a proteinaceous complex that is essential for proper chromosome segregation. As a core member of the inner kinetochore, defects of each subunit in the CENP-H/I/K complex cause dysfunction of kinetochore that leads to chromosome mis-segregation and cell death. However, how the CENP-H/I/K complex assembles and promotes kinetochore function are poorly understood. We here determined the crystal structures of CENP-I N-terminus alone from Chaetomium thermophilum and its complex with CENP-H/K from Thielavia terrestris, and verified the identified interactions. The structures and biochemical analyses show that CENP-H and CENP-K form a heterodimer through both N- and C-terminal interactions. CENP-I integrates into the CENP-H/K complex by binding to the C-terminus of CENP-H, leading to formation of the ternary complex in which CENP-H is sandwiched between CENP-K and CENP-I. Our sequence comparisons and mutational analyses showed that this architecture of the CENP-H/I/K complex is conserved in human. Mutating the binding interfaces of CENP-H for either CENP-K or CENP-I significantly reduced their localizations at centromeres and induced massive chromosome alignment defects during mitosis, suggesting that the identified interactions are critical for CENP-H/I/K complex assembly at the centromere and kinetochore function. Altogether, our findings unveil the evolutionarily conserved assembly mechanism of the CENP-H/I/K complex that is critical for proper chromosome alignment.
Organizational Affiliation:
Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan 430074, China.