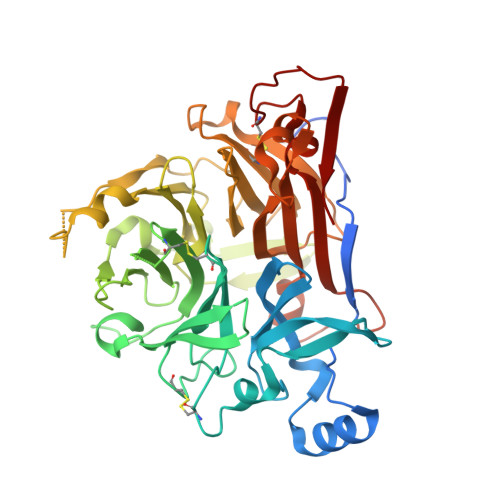
Architecture of the native major royal jelly protein 1 oligomer.
Tian, W., Li, M., Guo, H., Peng, W., Xue, X., Hu, Y., Liu, Y., Zhao, Y., Fang, X., Wang, K., Li, X., Tong, Y., Conlon, M.A., Wu, W., Ren, F., Chen, Z.(2018) Nat Commun 9: 3373-3373
- PubMed: 30135511
- DOI: https://doi.org/10.1038/s41467-018-05619-1
- Primary Citation of Related Structures:
5YYL - PubMed Abstract:
Honeybee caste development is nutritionally regulated by royal jelly (RJ). Major royal jelly protein 1 (MRJP1), the most abundant glycoprotein among soluble royal jelly proteins, plays pivotal roles in honeybee nutrition and larvae development, and exhibits broad pharmacological activities in humans. However, its structure has long remained unknown. Herein, we identify and report a 16-molecule architecture of native MRJP1 oligomer containing four MRJP1, four apisimin, and eight unanticipated 24-methylenecholesterol molecules at 2.65 Å resolution. MRJP1 has a unique six-bladed β-propeller fold with three disulfide bonds, and it interacts with apisimin mainly by hydrophobic interaction. Every four 24-methylenecholesterol molecules are packaged by two MRJP1 and two apisimin molecules. This assembly dimerizes to form an H-shaped MRJP1 4 -apisimin 4 -24-methylenecholesterol 8 complex via apisimin in a conserved and pH-dependent fashion. Our findings offer a structural basis for understanding the pharmacological effects of MRJPs and 24-methylenecholesterol, and provide insights into their unique physiological roles in bees.
Organizational Affiliation:
Beijing Advanced Innovation Center for Food Nutrition and Human Health, State Key Laboratory of Agrobiotechnology, China Agricultural University, Beijing, 100193, China.