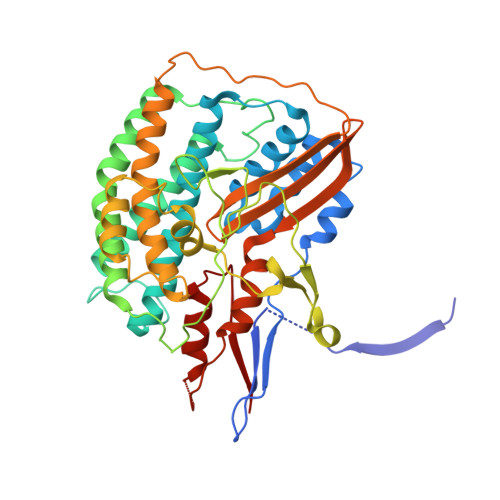
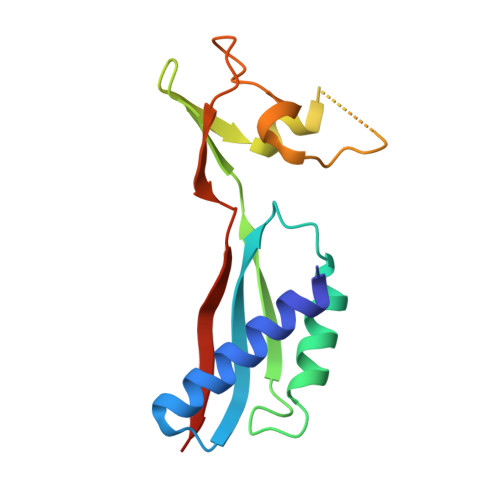
Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Kwon, S., Watanabe, S., Nishitani, Y., Kawashima, T., Kanai, T., Atomi, H., Miki, K.(2018) Proc Natl Acad Sci U S A 115: 7045-7050
- PubMed: 29915046
- DOI: https://doi.org/10.1073/pnas.1801955115
- Primary Citation of Related Structures:
5YXY, 5YY0 - PubMed Abstract:
Ni-Fe clusters are inserted into the large subunit of [NiFe] hydrogenases by maturation proteins such as the Ni chaperone HypA via an unknown mechanism. We determined crystal structures of an immature large subunit HyhL complexed with HypA from Thermococcus kodakarensis Structure analysis revealed that the N-terminal region of HyhL extends outwards and interacts with the Ni-binding domain of HypA. Intriguingly, the C-terminal extension of immature HyhL, which is cleaved in the mature form, adopts a β-strand adjacent to its N-terminal β-strands. The position of the C-terminal extension corresponds to that of the N-terminal extension of a mature large subunit, preventing the access of endopeptidases to the cleavage site of HyhL. These findings suggest that Ni insertion into the active site induces spatial rearrangement of both the N- and C-terminal tails of HyhL, which function as a key checkpoint for the completion of the Ni-Fe cluster assembly.
Organizational Affiliation:
Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, 606-8502 Kyoto, Japan.