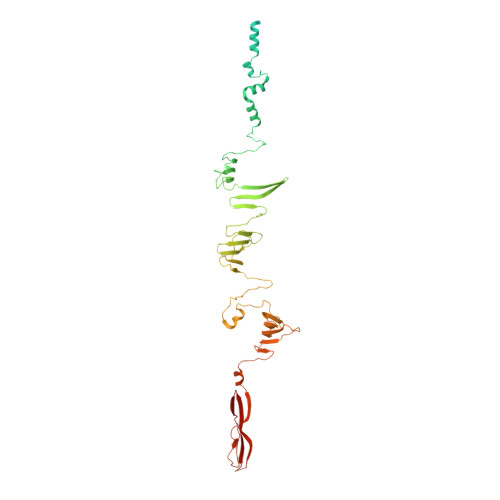
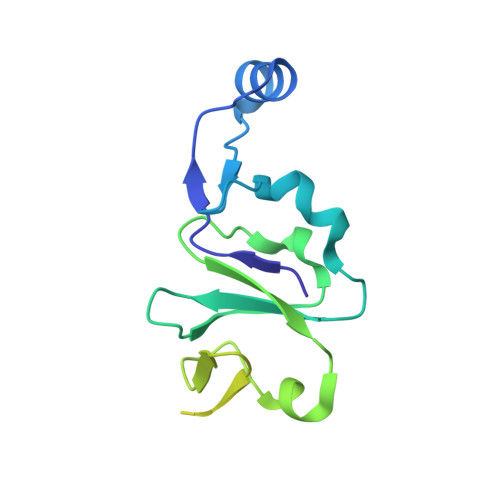
Phage tail fibre assembly proteins employ a modular structure to drive the correct folding of diverse fibres.
North, O.I., Sakai, K., Yamashita, E., Nakagawa, A., Iwazaki, T., Buttner, C.R., Takeda, S., Davidson, A.R.(2019) Nat Microbiol 4: 1645-1653
- PubMed: 31209305
- DOI: https://doi.org/10.1038/s41564-019-0477-7
- Primary Citation of Related Structures:
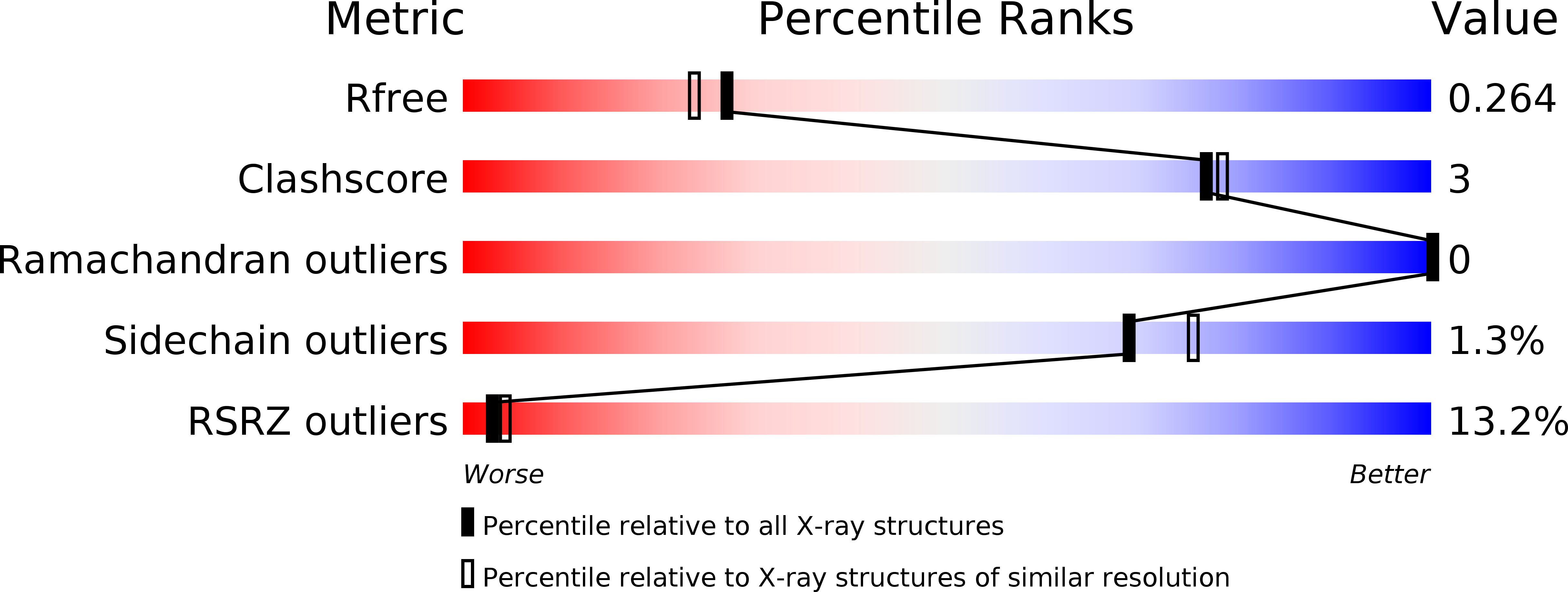
5YVQ - PubMed Abstract:
Phage tail fibres are elongated protein assemblies capable of specific recognition of bacterial surfaces during the first step of viral infection 1-4 . The folding of these complex trimeric structures often requires a phage-encoded tail fibre assembly (Tfa) protein 5-7 . Despite the wide occurrence of Tfa proteins, their functional mechanism has not been elucidated. Here, we investigate the tail fibre and Tfa of Escherichia coli phage Mu. We demonstrate that Tfa forms a stable complex with the tail fibre, and present a 2.1 Å resolution X-ray crystal structure of this complex. We find that Tfa proteins are comprised of two domains: a non-conserved N-terminal domain that binds to the C-terminal region of the fibre and a conserved C-terminal domain that probably mediates fibre oligomerization and assembly. Tfa forms rapidly exchanging multimers on its own, but not a stable trimer, implying that Tfa does not specify the trimeric state of the fibre. We propose that the key conserved role of Tfa is to ensure that fibre assembly and multimerization initiates at the C terminus, ensuring that the intertwined and repetitive structural elements of fibres come together in the correct sequence. The universal importance of correctly aligning the C termini of phage fibres is highlighted by our work.
Organizational Affiliation:
Department of Biochemistry, University of Toronto, Toronto, Ontario, Canada.