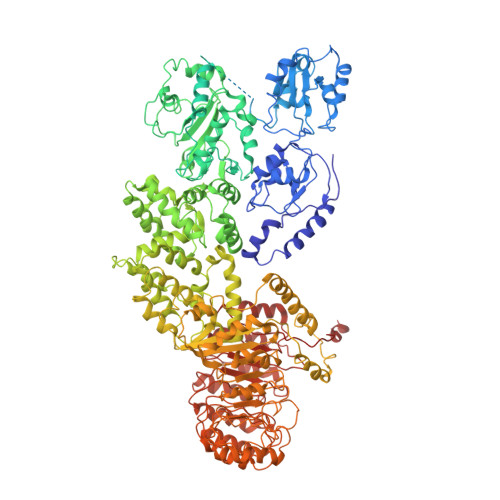
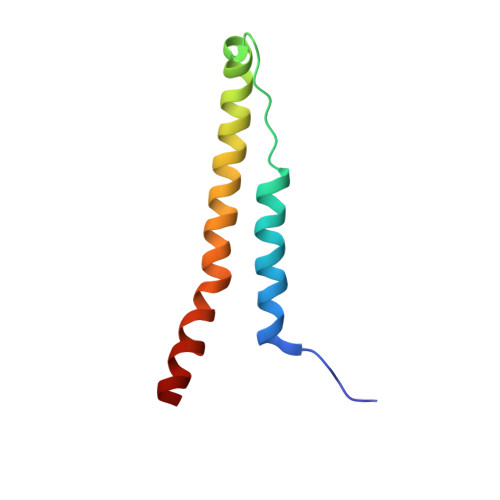
Structural basis for specific flagellin recognition by the NLR protein NAIP5.
Yang, X., Yang, F., Wang, W., Lin, G., Hu, Z., Han, Z., Qi, Y., Zhang, L., Wang, J., Sui, S.F., Chai, J.(2018) Cell Res 28: 35-47
- PubMed: 29182158
- DOI: https://doi.org/10.1038/cr.2017.148
- Primary Citation of Related Structures:
5YUD - PubMed Abstract:
The nucleotide-binding domain- and leucine-rich repeat (LRR)-containing proteins (NLRs) function as intracellular immune receptors to detect the presence of pathogen- or host-derived signals. The mechanisms of how NLRs sense their ligands remain elusive. Here we report the structure of a bacterial flagellin derivative in complex with the NLR proteins NAIP5 and NLRC4 determined by cryo-electron microscopy at 4.28 Å resolution. The structure revealed that the flagellin derivative forms two parallel helices interacting with multiple domains including BIR1 and LRR of NAIP5. Binding to NAIP5 results in a nearly complete burial of the flagellin derivative, thus stabilizing the active conformation of NAIP5. The extreme C-terminal side of the flagellin is anchored to a sterically constrained binding pocket of NAIP5, which likely acts as a structural determinant for discrimination of different bacterial flagellins by NAIP5, a notion further supported by biochemical data. Taken together, our results shed light on the molecular mechanisms underlying NLR ligand perception.
Organizational Affiliation:
Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing 100084, China.