C-terminal Redox Domain ofArabidopsisAPR1 is a Non-Canonical Thioredoxin Domain with Glutaredoxin Function.
Chen, F.F., Chien, C.Y., Cho, C.C., Chang, Y.Y., Hsu, C.H.(2019) Antioxidants (Basel) 8
- PubMed: 31597378
- DOI: https://doi.org/10.3390/antiox8100461
- Primary Citation of Related Structures:
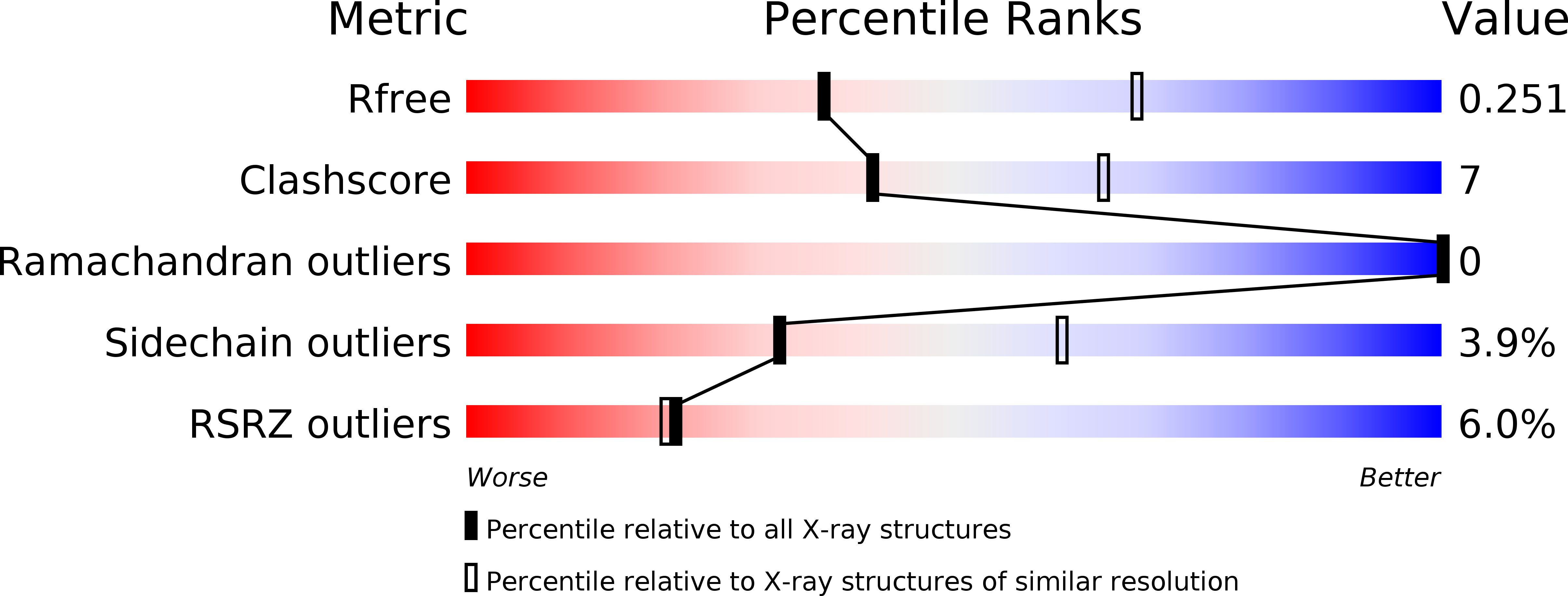
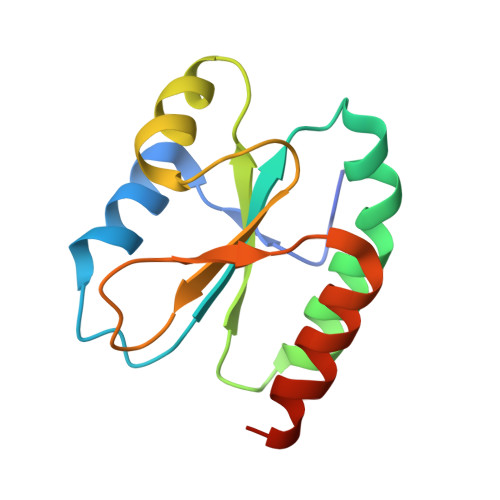
5YRY - PubMed Abstract:
Sulfur is an essential nutrient that can be converted into utilizable metabolic forms to produce sulfur-containing metabolites in plant. Adenosine 5'-phosphosulfate (APS) reductase (APR) plays a vital role in catalyzing the reduction of activated sulfate to sulfite, which requires glutathione. Previous studies have shown that the C-terminal domain of APR acts as a glutathione-dependent reductase. The crystal structure of the C-terminal redox domain of Arabidopsis APR1 (AtAPR1) shows a conserved α/β thioredoxin fold, but not a glutaredoxin fold. Further biochemical studies of the redox domain from AtAPR1 provided evidence to support the structural observation. Collectively, our results provide structural and biochemical information to explain how the thioredoxin fold exerts the glutaredoxin function in APR.
Organizational Affiliation:
Department of Agricultural Chemistry, National Taiwan University, Taipei 10617, Taiwan. fun_0108@hotmail.com.