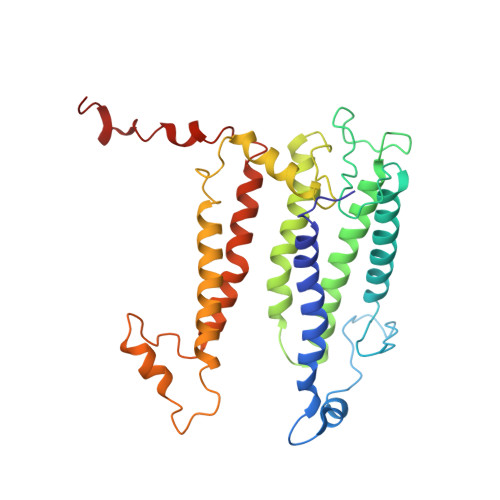
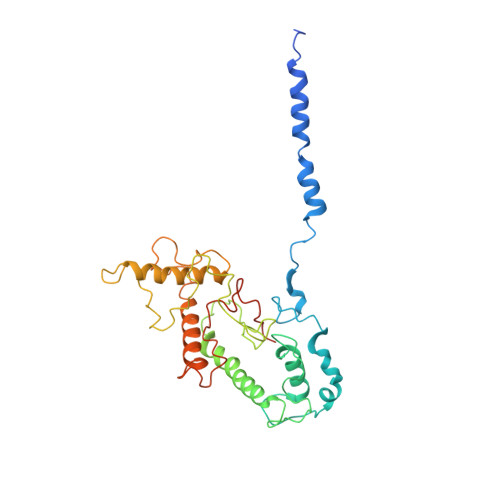
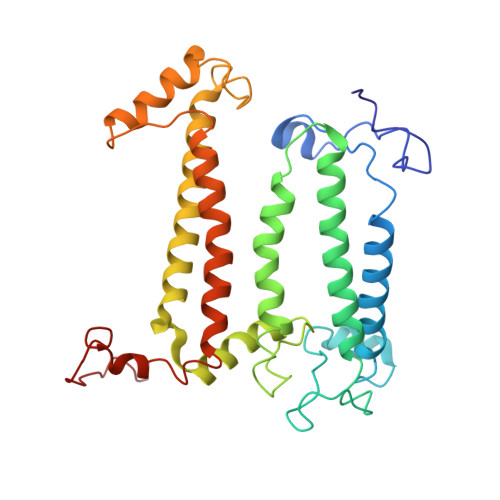
Cryo-EM structure of the RC-LH core complex from an early branching photosynthetic prokaryote.
Xin, Y.Y., Shi, Y., Niu, T.X., Wang, Q.Q., Niu, W.Q., Huang, X.J., Ding, W., Yang, L., Blankenship, R.E., Xu, X.L., Sun, F.(2018) Nat Commun 9: 1568-1568
- PubMed: 29674684
- DOI: https://doi.org/10.1038/s41467-018-03881-x
- Primary Citation of Related Structures:
5YQ7 - PubMed Abstract:
Photosynthetic prokaryotes evolved diverse light-harvesting (LH) antennas to absorb sunlight and transfer energy to reaction centers (RC). The filamentous anoxygenic phototrophs (FAPs) are important early branching photosynthetic bacteria in understanding the origin and evolution of photosynthesis. How their photosynthetic machinery assembles for efficient energy transfer is yet to be elucidated. Here, we report the 4.1 Å structure of photosynthetic core complex from Roseiflexus castenholzii by cryo-electron microscopy. The RC-LH complex has a tetra-heme cytochrome c bound RC encompassed by an elliptical LH ring that is assembled from 15 LHαβ subunits. An N-terminal transmembrane helix of cytochrome c inserts into the LH ring, not only yielding a tightly bound cytochrome c for rapid electron transfer, but also opening a slit in the LH ring, which is further flanked by a transmembrane helix from a newly discovered subunit X. These structural features suggest an unusual quinone exchange model of prokaryotic photosynthetic machinery.
Organizational Affiliation:
Hangzhou Normal University, 2318 Yuhangtang Road, Cangqian, Yuhang District, Hangzhou, 311121, Zhejiang Province, China.