Comparative structural and enzymatic studies on Salmonella typhimurium diaminopropionate ammonia lyase reveal its unique features
Deka, G., Bisht, S., Savithri, H.S., Murthy, M.R.N.(2018) J Struct Biol 202: 118-128
- PubMed: 29294403
- DOI: https://doi.org/10.1016/j.jsb.2017.12.012
- Primary Citation of Related Structures:
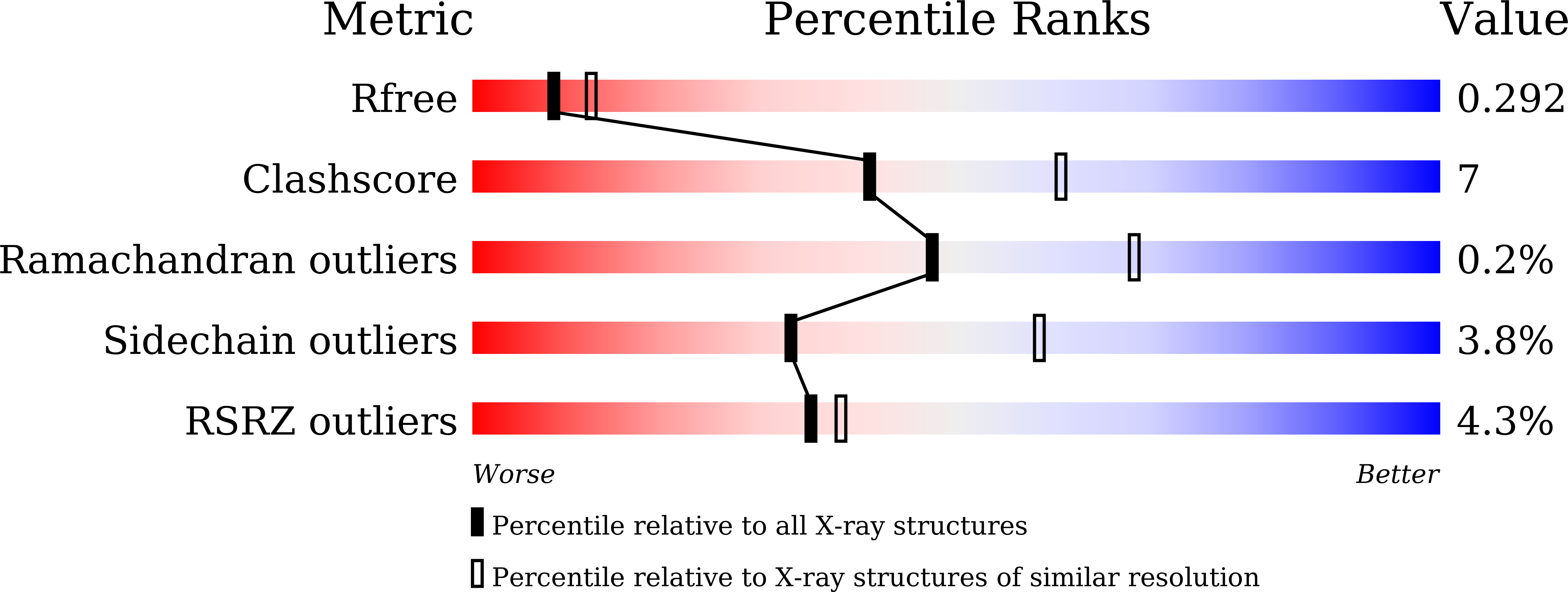
5YGR - PubMed Abstract:
Cellular metabolism of amino acids is controlled by a large number of pyridoxal 5'-phosphate (PLP) dependent enzymes. Diaminopropionate ammonia lyase (DAPAL), a fold type II PLP-dependent enzyme, degrades both the D and L forms of diaminopropionic acid (DAP) to pyruvate and ammonia. Earlier studies on the Escherichia coli DAPAL (EcDAPAL) had suggested that a disulfide bond located close to the active site may be crucial for maintaining the geometry of the substrate entry channel and the active site. In order to obtain further insights into the catalytic properties of DAPAL, structural and functional studies on Salmonella typhimurium DAPAL (StDAPAL) were initiated. The three-dimensional X-ray crystal structure of StDAPAL was determined at 2.5 Å resolution. As expected, the polypeptide fold and dimeric organization of StDAPAL is similar to those of EcDAPAL. A phosphate group was located in the active site of StDAPAL and expulsion of this phosphate is probably essential to bring Asp125 to a conformation suitable for proton abstraction from the substrate (D-DAP). The unique disulfide bond of EcDAPAL was absent in StDAPAL, although the enzyme displayed comparable catalytic activity. Site directed mutagenesis of the cysteine residues involved in disulfide bond formation in EcDAPAL followed by functional and biophysical studies further confirmed that the disulfide bond is not necessary either for substrate binding or for catalysis. The activity of StDAPAL but not EcDAPAL was enhanced by monovalent cations suggesting subtle differences in the active site geometries of these two closely related enzymes.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore 560012, India.