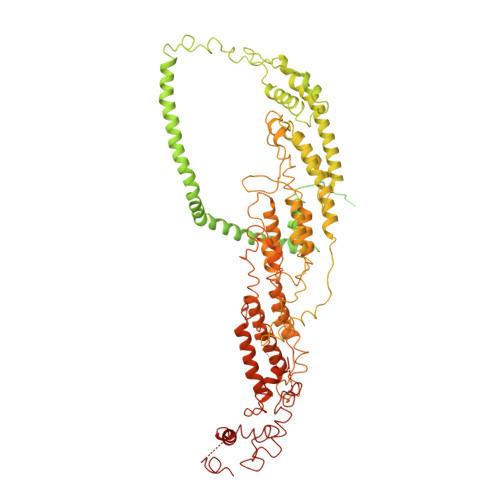
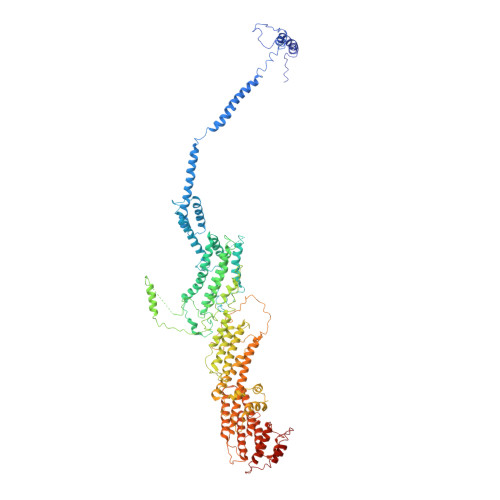
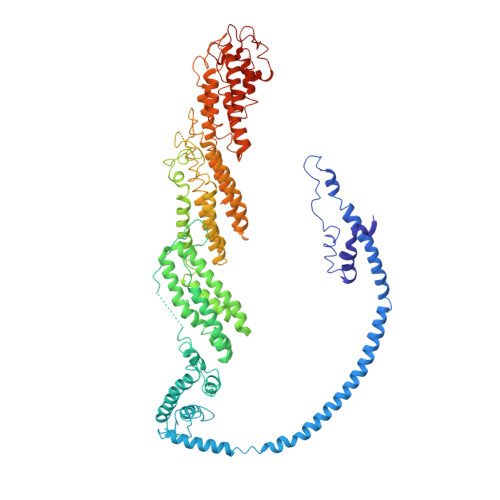
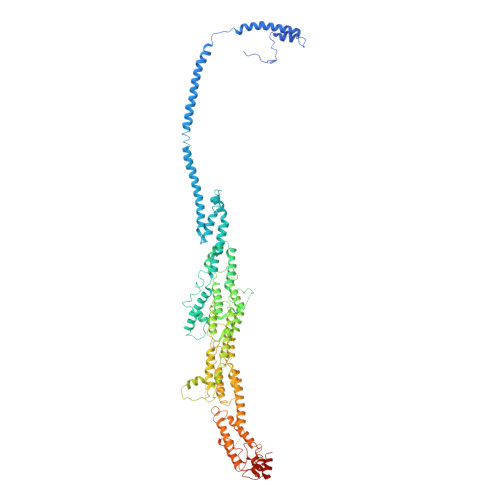
Cryo-EM structure of the exocyst complex
Mei, K., Li, Y., Wang, S., Shao, G., Wang, J., Ding, Y., Luo, G., Yue, P., Liu, J.J., Wang, X., Dong, M.Q., Wang, H.W., Guo, W.(2018) Nat Struct Mol Biol 25: 139-146
- PubMed: 29335562
- DOI: https://doi.org/10.1038/s41594-017-0016-2
- Primary Citation of Related Structures:
5YFP - PubMed Abstract:
The exocyst is an evolutionarily conserved octameric protein complex that mediates the tethering of post-Golgi secretory vesicles to the plasma membrane during exocytosis and is implicated in many cellular processes such as cell polarization, cytokinesis, ciliogenesis and tumor invasion. Using cryo-EM and chemical cross-linking MS (CXMS), we solved the structure of the Saccharomyces cerevisiae exocyst complex at an average resolution of 4.4 Å. Our model revealed the architecture of the exocyst and led to the identification of the helical bundles that mediate the assembly of the complex at its core. Sequence analysis suggests that these regions are evolutionarily conserved across eukaryotic systems. Additional cell biological data suggest a mechanism for exocyst assembly that leads to vesicle tethering at the plasma membrane.
Organizational Affiliation:
Department of Biology, University of Pennsylvania, Philadelphia, PA, USA.