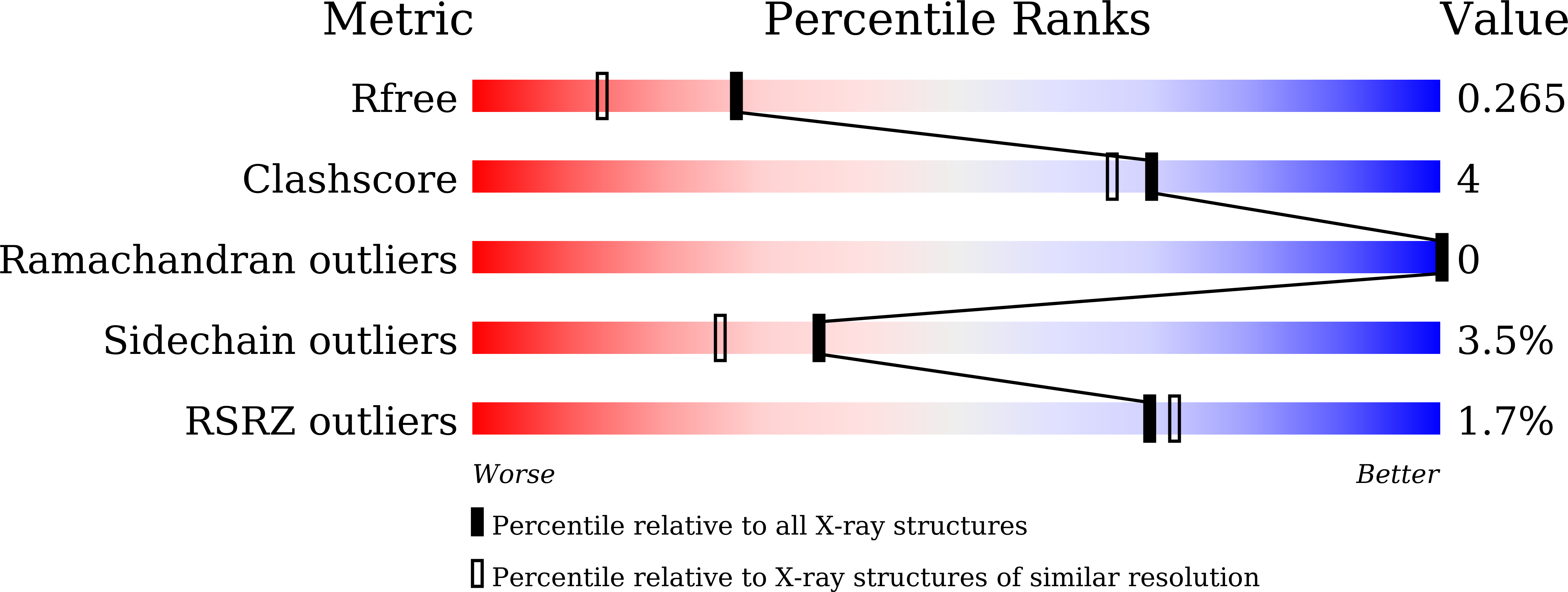
Characteristics of the essential pathogenicity factor Rv1828, a MerR family transcription regulator from Mycobacterium tuberculosis.
Singh, S., Sevalkar, R.R., Sarkar, D., Karthikeyan, S.(2018) FEBS J 285: 4424-4444
- PubMed: 30306715
- DOI: https://doi.org/10.1111/febs.14676
- Primary Citation of Related Structures:
5YDC, 5YDD - PubMed Abstract:
The gene Rv1828 in Mycobacterium tuberculosis is shown to be essential for the pathogen and encodes for an uncharacterized protein. In this study, we have carried out biochemical and structural characterization of Rv1828 at the molecular level to understand its mechanism of action. The Rv1828 is annotated as helix-turn-helix (HTH)-type MerR family transcription regulator based on its N-terminal amino acid sequence similarity. The MerR family protein binds to a specific DNA sequence in the spacer region between -35 and -10 elements of a promoter through its N-terminal domain (NTD) and acts as transcriptional repressor or activator depending on the absence or presence of effector that binds to its C-terminal domain (CTD). A characteristic feature of MerR family protein is its ability to bind to 19 ± 1 bp DNA sequence in the spacer region between -35 and -10 elements which is otherwise a suboptimal length for transcription initiation by RNA polymerase. Here, we show the Rv1828 through its NTD binds to a specific DNA sequence that exists on its own as well as in other promoter regions. Moreover, the crystal structure of CTD of Rv1828, determined by single-wavelength anomalous diffraction method, reveals a distinctive dimerization. The biochemical and structural analysis reveals that Rv1828 specifically binds to an everted repeat through its winged-HTH motif. Taken together, we demonstrate that the Rv1828 encodes for a MerR family transcription regulator.
Organizational Affiliation:
CSIR-Institute of Microbial Technology, Council of Scientific and Industrial Research, Chandigarh, India.