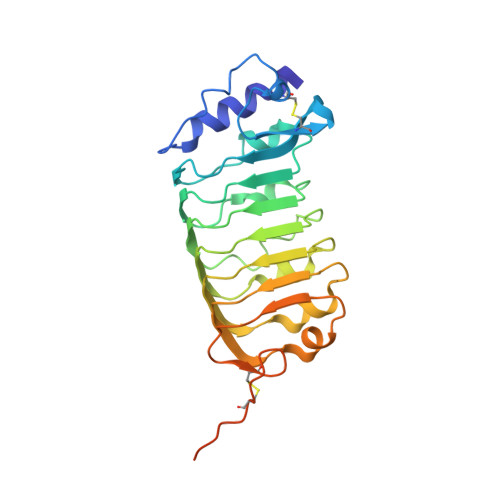
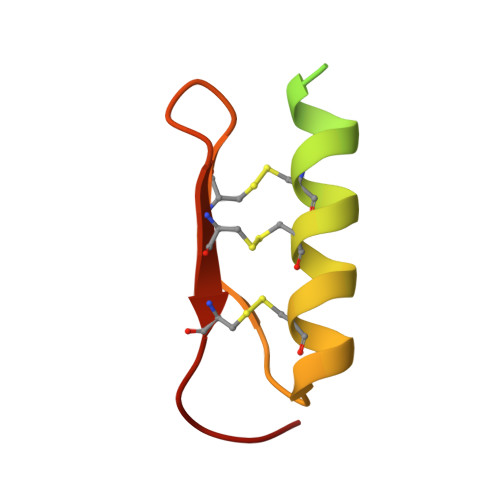
Structural basis for receptor recognition of pollen tube attraction peptides.
Zhang, X., Liu, W., Nagae, T.T., Takeuchi, H., Zhang, H., Han, Z., Higashiyama, T., Chai, J.(2017) Nat Commun 8: 1331-1331
- PubMed: 29109411
- DOI: https://doi.org/10.1038/s41467-017-01323-8
- Primary Citation of Related Structures:
5Y9W, 5YAH - PubMed Abstract:
Transportation of the immobile sperms directed by pollen tubes to the ovule-enclosed female gametophytes is important for plant sexual reproduction. The defensin-like (DEFL) cysteine-rich peptides (CRPs) LUREs play an essential role in pollen tube attraction to the ovule, though their receptors still remain controversial. Here we provide several lines of biochemical evidence showing that the extracellular domain of the leucine-rich repeat receptor kinase (LRR-RK) PRK6 from Arabidopsis thaliana directly interacts with AtLURE1 peptides. Structural study reveals that a C-terminal loop of the LRR domain (AtPRK6 LRR ) is responsible for recognition of AtLURE1.2, mediated by a set of residues largely conserved among PRK6 homologs from Arabidopsis lyrata and Capsella rubella, supported by in vitro mutagenesis and semi-in-vivo pollen tube growth assays. Our study provides evidence showing that PRK6 functions as a receptor of the LURE peptides in A. thaliana and reveals a unique ligand recognition mechanism of LRR-RKs.
Organizational Affiliation:
Ministry of Education Key Laboratory of Protein Science, Center for Structural Biology, School of Life Sciences, Tsinghua-Peking Joint Center for Life Sciences, Tsinghua University, 100084, Beijing, China.