Structural Insights into Yeast Telomerase Recruitment to Telomeres
Chen, H., Xue, J., Churikov, D., Hass, E.P., Shi, S., Lemon, L.D., Luciano, P., Bertuch, A.A., Zappulla, D.C., Geli, V., Wu, J., Lei, M.(2018) Cell 172: 331-343.e13
- PubMed: 29290466
- DOI: https://doi.org/10.1016/j.cell.2017.12.008
- Primary Citation of Related Structures:
5Y58, 5Y59, 5Y5A - PubMed Abstract:
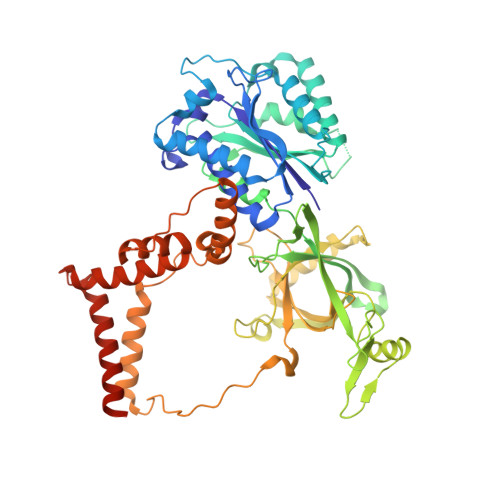
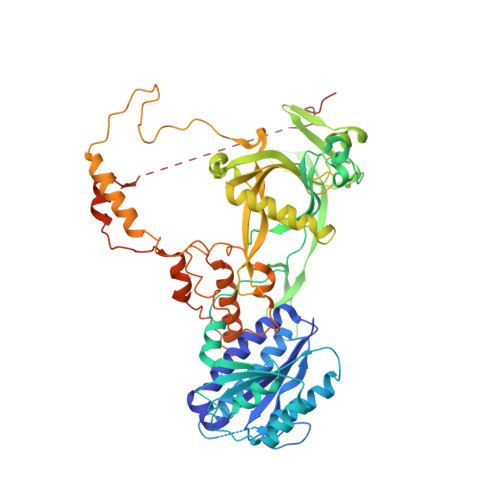
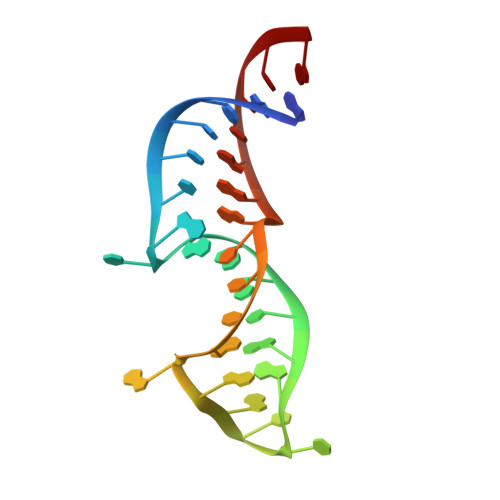
Telomerase maintains chromosome ends from humans to yeasts. Recruitment of yeast telomerase to telomeres occurs through its Ku and Est1 subunits via independent interactions with telomerase RNA (TLC1) and telomeric proteins Sir4 and Cdc13, respectively. However, the structures of the molecules comprising these telomerase-recruiting pathways remain unknown. Here, we report crystal structures of the Ku heterodimer and Est1 complexed with their key binding partners. Two major findings are as follows: (1) Ku specifically binds to telomerase RNA in a distinct, yet related, manner to how it binds DNA; and (2) Est1 employs two separate pockets to bind distinct motifs of Cdc13. The N-terminal Cdc13-binding site of Est1 cooperates with the TLC1-Ku-Sir4 pathway for telomerase recruitment, whereas the C-terminal interface is dispensable for binding Est1 in vitro yet is nevertheless essential for telomere maintenance in vivo. Overall, our results integrate previous models and provide fundamentally valuable structural information regarding telomere biology.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 201210 Shanghai, China.