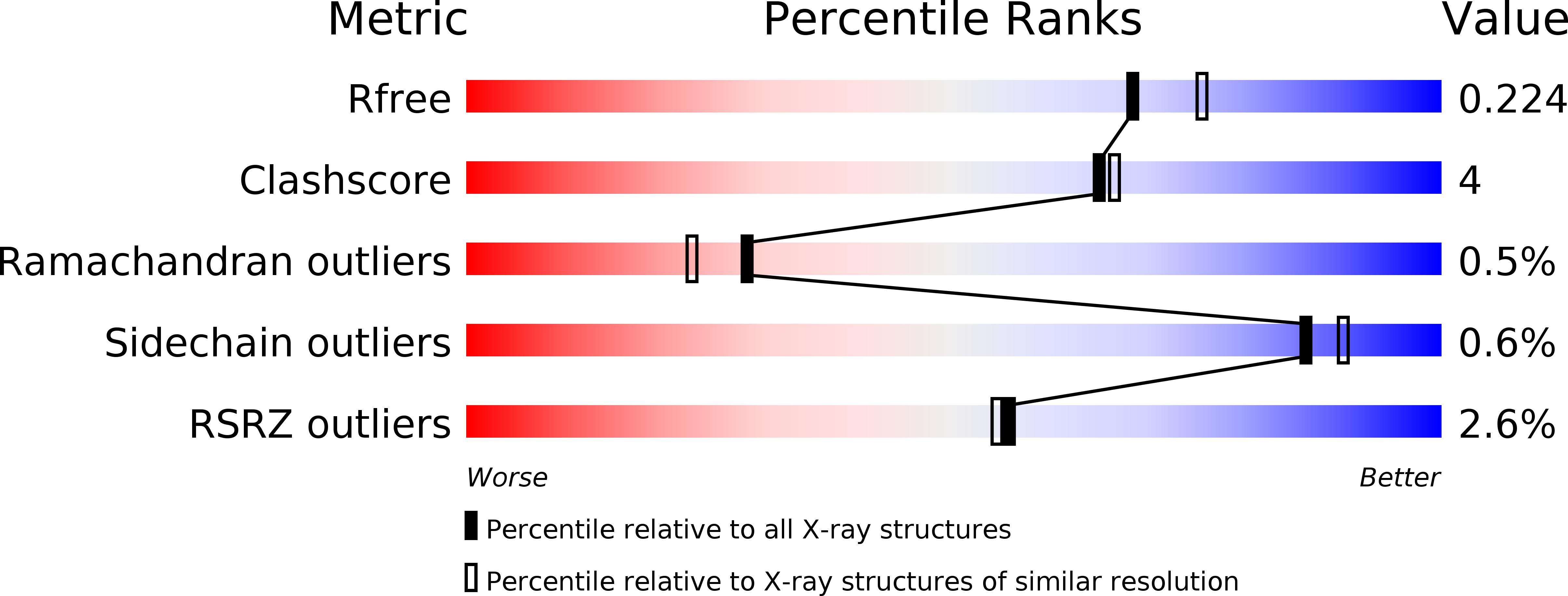
Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex.
He, H., Li, Y., Dong, Q., Chang, A.Y., Gao, F., Chi, Z., Su, M., Zhang, F., Ban, H., Martienssen, R., Chen, Y.H., Li, F.(2017) Proc Natl Acad Sci U S A 114: 12524-12529
- PubMed: 29109278
- DOI: https://doi.org/10.1073/pnas.1712961114
- Primary Citation of Related Structures:
5Y26, 5Y27 - PubMed Abstract:
During DNA replication, chromatin is disrupted ahead of the replication fork, and epigenetic information must be restored behind the fork. How epigenetic marks are inherited through DNA replication remains poorly understood. Histone H3 lysine 9 (H3K9) methylation and histone hypoacetylation are conserved hallmarks of heterochromatin. We previously showed that the inheritance of H3K9 methylation during DNA replication depends on the catalytic subunit of DNA polymerase epsilon, Cdc20. Here we show that the histone-fold subunit of Pol epsilon, Dpb4, interacts an uncharacterized small histone-fold protein, SPCC16C4.22, to form a heterodimer in fission yeast. We demonstrate that SPCC16C4.22 is nonessential for viability and corresponds to the true ortholog of Dpb3. We further show that the Dpb3-Dpb4 dimer associates with histone deacetylases, chromatin remodelers, and histones and plays a crucial role in the inheritance of histone hypoacetylation in heterochromatin. We solve the 1.9-Å crystal structure of Dpb3-Dpb4 and reveal that they form the H2A-H2B-like dimer. Disruption of Dpb3-Dpb4 dimerization results in loss of heterochromatin silencing. Our findings reveal a link between histone deacetylation and H3K9 methylation and suggest a mechanism for how two processes are coordinated during replication. We propose that the Dpb3-Dpb4 heterodimer together with Cdc20 serves as a platform for the recruitment of chromatin modifiers and remodelers that mediate heterochromatin assembly during DNA replication, and ensure the faithful inheritance of epigenetic marks in heterochromatin.
Organizational Affiliation:
Department of Biology, New York University, New York, NY 10003-6688.