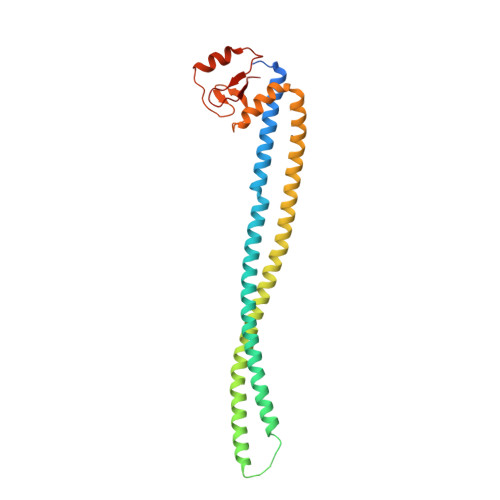
Crystal structure of the MSMEG_4306 gene product from Mycobacterium smegmatis
Kumar, A., Karthikeyan, S.(2018) Acta Crystallogr F Struct Biol Commun 74: 166-173
- PubMed: 29497021
- DOI: https://doi.org/10.1107/S2053230X18002236
- Primary Citation of Related Structures:
5Y05, 5Y06 - PubMed Abstract:
The MSMEG_4306 gene from Mycobacterium smegmatis encodes a protein of unknown function with 242 amino-acid residues that contains a conserved zinc-ribbon domain at its C-terminus. Here, the crystal structure of MSMEG_4306 determined by the single-wavelength anomalous dispersion method using just one zinc ion co-purified with the protein is reported. The crystal structure of MSMEG_4306 shows a coiled-coil helix domain in the N-terminal region and a zinc-ribbon domain in the C-terminal region. A structural similarity search against the Protein Data Bank using MSMEG_4306 as a query revealed two similar structures, namely CT398 from Chlamydia trachomatis and HP0958 from Helicobacter pylori, although they share only ∼15% sequence identity with MSMEG_4306. Based on comparative analysis, it is predicted that MSMEG_4306 may be involved in secretion systems, possibly by interacting with multiple proteins or nucleic acids.
Organizational Affiliation:
CSIR - Institute of Microbial Technology, Council of Scientific and Industrial Research (CSIR), Sector 39A, Chandigarh 160 036, India.