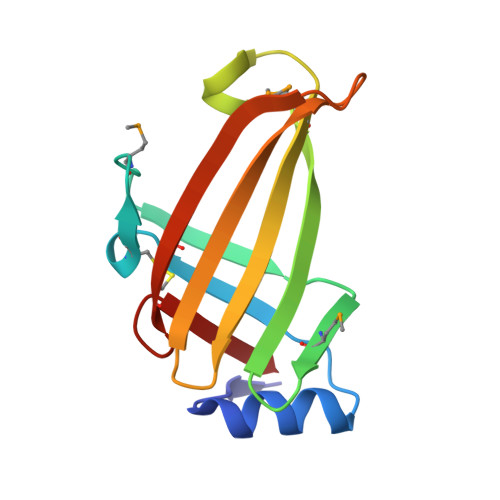
Crystal structure of lpg1832, a VirK family protein from Legionella pneumophila, reveals a novel fold for bacterial VirK proteins
Zhang, N., Yin, S., Liu, S., Sun, A., Zhou, M., Gong, X., Ge, H.(2017) FEBS Lett 591: 2929-2935
- PubMed: 28771688
- DOI: https://doi.org/10.1002/1873-3468.12773
- Primary Citation of Related Structures:
5XTA - PubMed Abstract:
VirK family [Pfam06903] consists of 14 bacterial VirK proteins of around 145 residues in length. The function of this family is unknown. Herein, using single-wavelength anomalous diffraction, we determined the crystal structure of lpg1832, a VirK family protein from Legionella pneumophila, at 2.0 Å resolution. This is the first structural determination of a VirK domain-containing protein. Lpg1832 is a type II secretion system-dependent extracellular protein that folds into a novel barrel-shaped structure. It is found to adopt a quaternary assembly comprising a homotetramer. The three-dimensional structure of lpg1832 provides the first structural information pertaining to the VirK family and allows us to possibly identify its functionally important regions.
Organizational Affiliation:
Institute of Health Sciences, School of Life Sciences, Anhui University, Hefei, China.