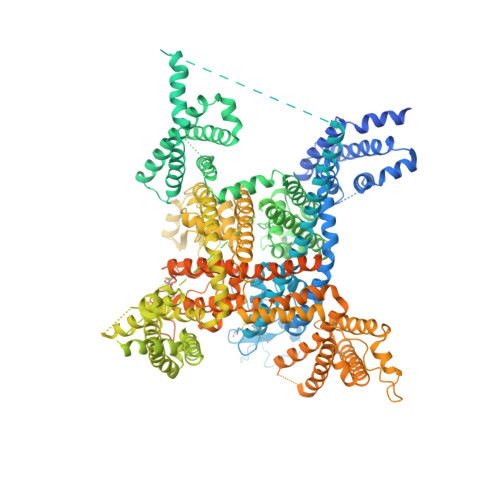
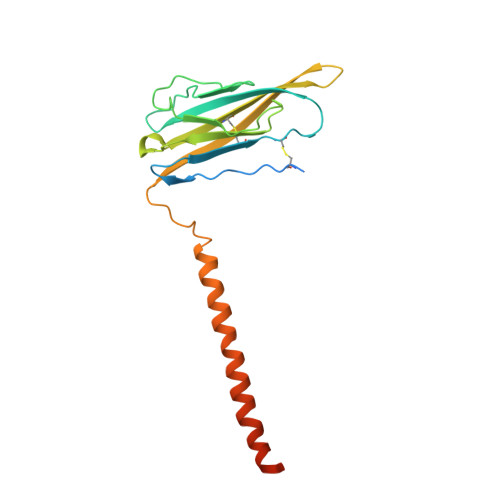
Structure of the Nav1.4-beta 1 Complex from Electric Eel.
Yan, Z., Zhou, Q., Wang, L., Wu, J., Zhao, Y., Huang, G., Peng, W., Shen, H., Lei, J., Yan, N.(2017) Cell 170: 470-482.e11
- PubMed: 28735751
- DOI: https://doi.org/10.1016/j.cell.2017.06.039
- Primary Citation of Related Structures:
5XSY - PubMed Abstract:
Voltage-gated sodium (Na v ) channels initiate and propagate action potentials. Here, we present the cryo-EM structure of EeNa v 1.4, the Na v channel from electric eel, in complex with the β1 subunit at 4.0 Å resolution. The immunoglobulin domain of β1 docks onto the extracellular L5 I and L6 IV loops of EeNa v 1.4 via extensive polar interactions, and the single transmembrane helix interacts with the third voltage-sensing domain (VSD III ). The VSDs exhibit "up" conformations, while the intracellular gate of the pore domain is kept open by a digitonin-like molecule. Structural comparison with closed Na v PaS shows that the outward transfer of gating charges is coupled to the iris-like pore domain dilation through intricate force transmissions involving multiple channel segments. The IFM fast inactivation motif on the III-IV linker is plugged into the corner enclosed by the outer S4-S5 and inner S6 segments in repeats III and IV, suggesting a potential allosteric blocking mechanism for fast inactivation.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences and School of Medicine, Tsinghua University, Beijing, China; Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing, China.