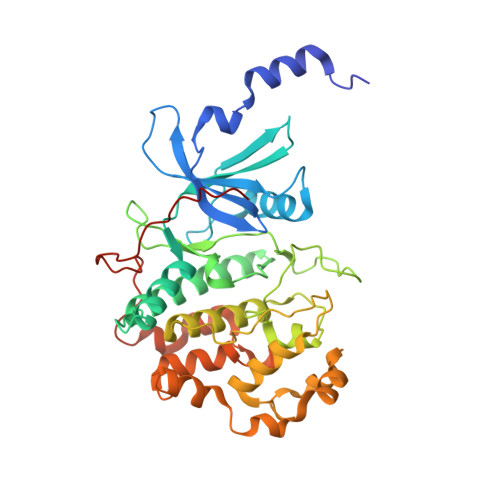
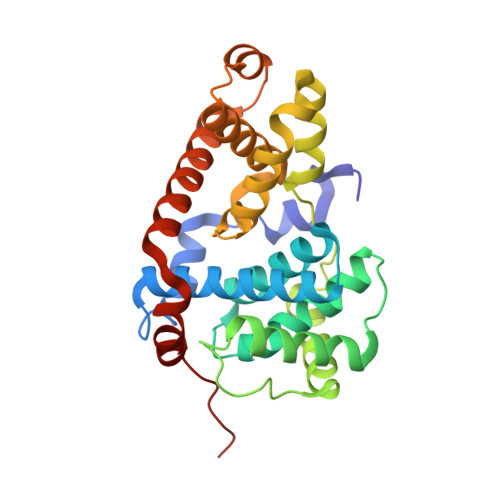
Discovery of potent and selective CDK8 inhibitors through FBDD approach
Han, X., Jiang, M., Zhou, C., Zhou, Z., Xu, Z., Wang, L., Mayweg, A.V., Niu, R., Jin, T.G., Yang, S.(2017) Bioorg Med Chem Lett 27: 4488-4492
- PubMed: 28802632
- DOI: https://doi.org/10.1016/j.bmcl.2017.07.080
- Primary Citation of Related Structures:
5XQX, 5XS2 - PubMed Abstract:
A fragment library screen was carried out to identify starting points for novel CDK8 inhibitors. Optimization of a fragment hit guided by co-crystal structures led to identification of a novel series of potent CDK8 inhibitors which are highly ligand efficient, kinase selective and cellular active. Compound 16 was progressed to a mouse pharmacokinetic study and showed good oral bioavailability.
Organizational Affiliation:
Medicinal Chemistry, Roche Innovation Center Shanghai, Bldg 5, 720 Cailun Road, Shanghai 201203, China. Electronic address: cyrus.han@roche.com.