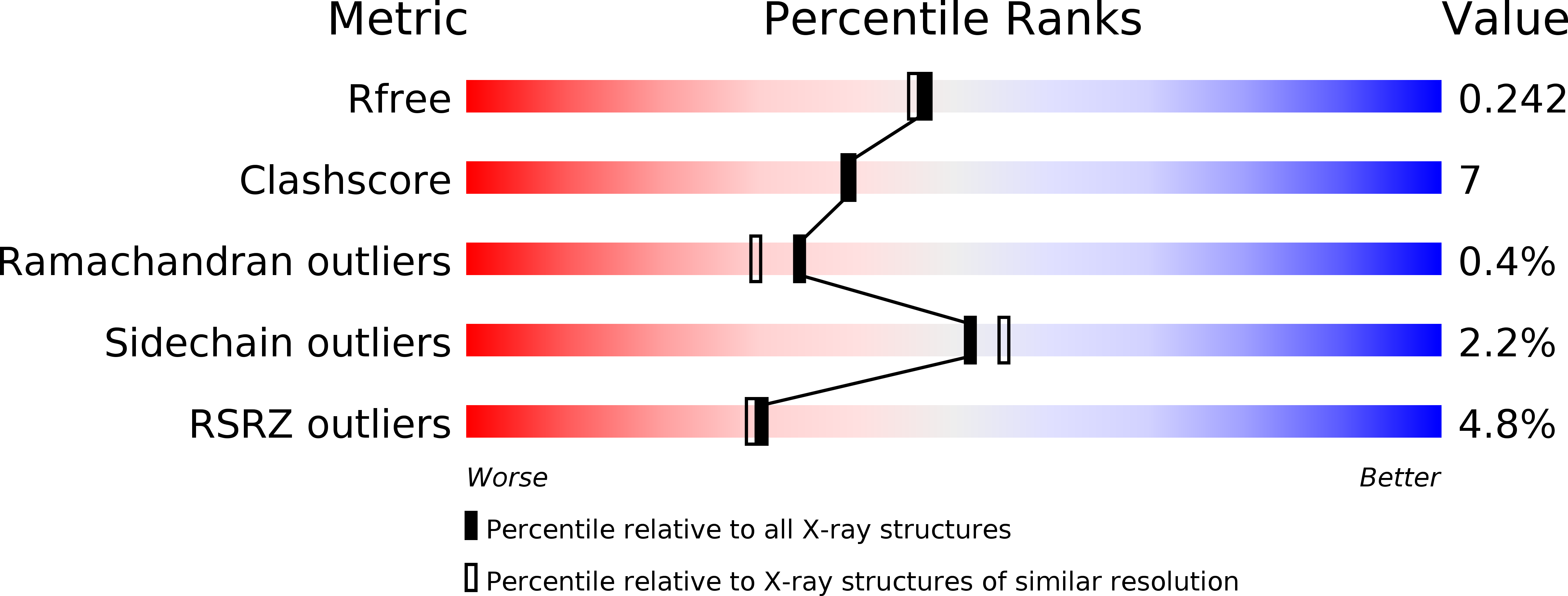
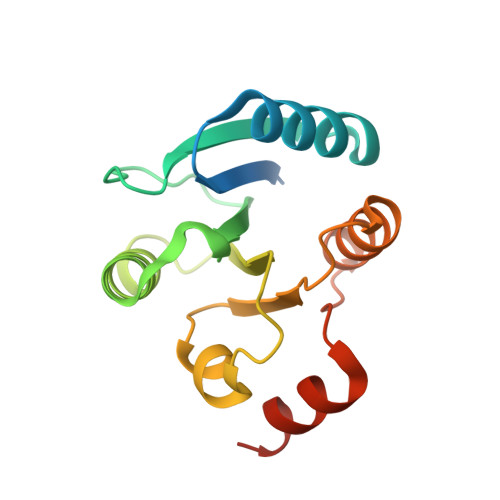
Crystal structure of master biofilm regulator CsgD regulatory domain reveals an atypical receiver domain.
Wen, Y., Ouyang, Z., Devreese, B., He, W., Shao, Y., Lu, W., Zheng, F.(2017) Protein Sci 26: 2073-2082
- PubMed: 28758290
- DOI: https://doi.org/10.1002/pro.3245
- Primary Citation of Related Structures:
5XP0 - PubMed Abstract:
The master regulator CsgD switches planktonic growth to biofilm formation by activating synthesis of curli fimbriae and cellulose in Enterobacteriaceae. CsgD was classified to be the LuxR response regulatory family, while its cognate sensor histidine kinase has not been identified yet. CsgD consists of a C-terminal DNA binding domain and an N-terminal regulatory domain that provokes the upstream signal transduction to further modulate its function. We provide the crystal structure of Salmonella Typhimurium CsgD regulatory domain, which reveals an atypical β5α5 response regulatory receiver domain folding with the α2 helix representing as a disorder loop compared to the LuxR/FixJ canonical response regulator, and the structure indicated a noteworthy α5 helix similar to the non-canonical master regulator VpsT receiver domain α6. CsgD regulatory domain assembles with two dimerization interfaces mainly through α1 and α5, which has shown similarity to the c-di-GMP independent and stabilized dimerization interface of VpsT from Vibrio cholerae respectively. The potential phosphorylation site D59 is directly involved in the interaction of interfaces I and mutagenesis studies indicated that both dimerization interfaces could be crucial for CsgD activity. The structure reveals important molecular details for the dimerization assembly of CsgD and will shed new insight into its regulation mechanism.
Organizational Affiliation:
Center for Translational Medicine, The Key Laboratory of Biomedical Information Engineering of Ministry of Education, School of Life Science and Technology, Xi'an Jiaotong University, Xi'an, China.