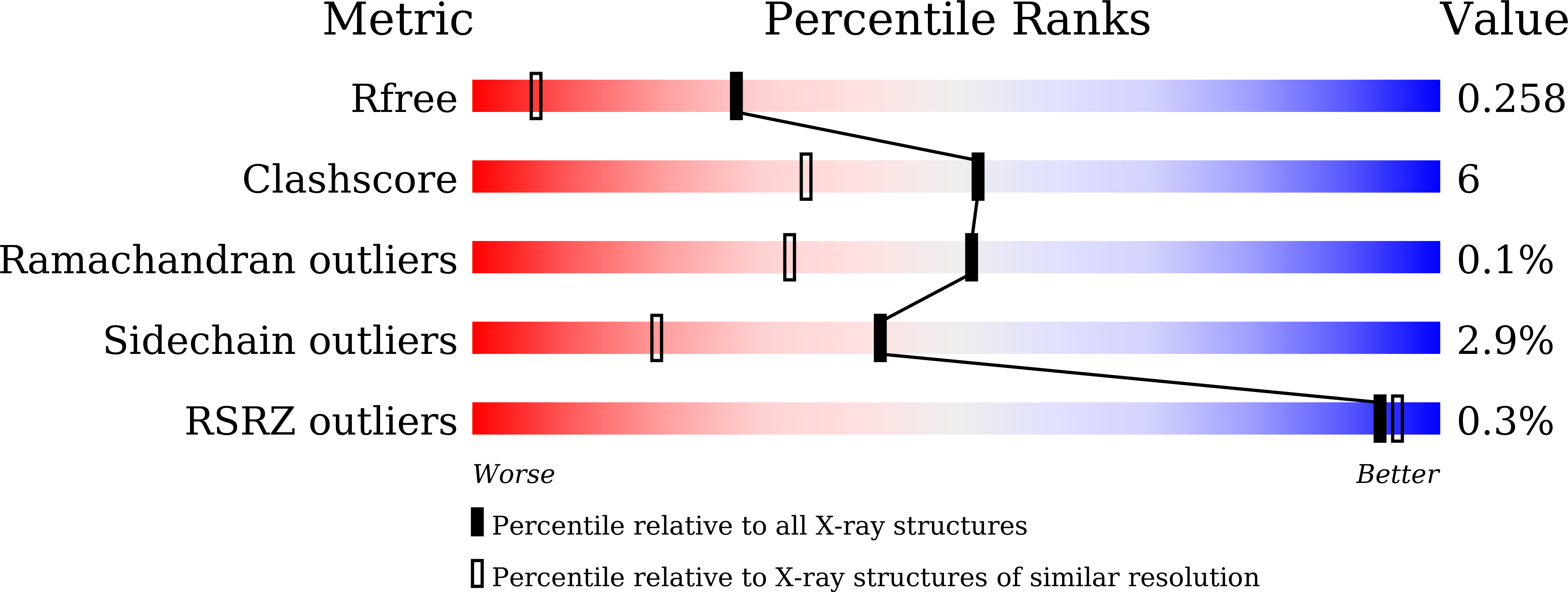
Improved thermostability of a metagenomic glucose-tolerant beta-glycosidase based on its X-ray crystal structure.
Matsuzawa, T., Watanabe, M., Yaoi, K.(2017) Appl Microbiol Biotechnol 101: 8353-8363
- PubMed: 29063172
- DOI: https://doi.org/10.1007/s00253-017-8525-9
- Primary Citation of Related Structures:
5XGZ - PubMed Abstract:
MeBglD2, a metagenomic β-glycosidase, is stimulated by various saccharides, including D-glucose, D-xylose, and maltose, and it promotes the enzymatic saccharification of plant biomass. To improve the thermostability of MeBglD2, its X-ray crystal structure was analyzed, and the amino acid residues responsible for its thermostability were identified using the structural information. Mutations in His8, Asn59, and Gly295 improved the thermostability of MeBglD2, and the combination of these mutations resulted in the highest thermostability. Compared with wild-type MeBglD2, thermostable MeBglD2 mutants promoted plant biomass saccharification using Trichoderma reesei cellulase. In addition to thermostability, the thermostable mutants exhibited higher tolerance to ethanol, dimethyl sulfoxide, and copper ions, indicating that the MeBglD2 mutants generated in this study were improved in their tolerance to not only high temperature but also to organic solvents and metal ions.
Organizational Affiliation:
Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Tsukuba Central 6, 1-1-1 Higashi, Tsukuba, Ibaraki, 305-8566, Japan. matsuzawa-tomohiko@aist.go.jp.