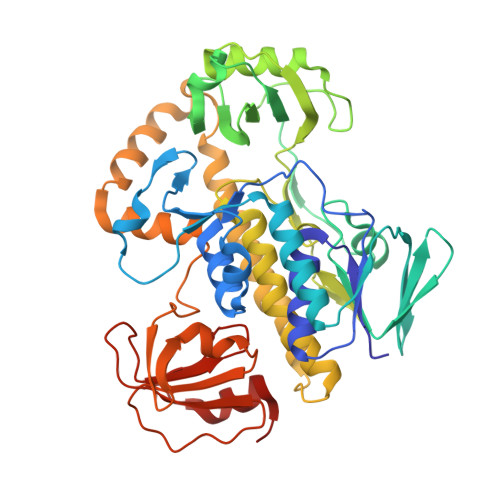
Structural Insights into a Flavin-Dependent [4 + 2] Cyclase that Catalyzes trans-Decalin Formation in Pyrroindomycin Biosynthesis.
Zheng, Q., Gong, Y., Guo, Y., Zhao, Z., Wu, Z., Zhou, Z., Chen, D., Pan, L., Liu, W.(2018) Cell Chem Biol 25: 718-727.e3
- PubMed: 29657086
- DOI: https://doi.org/10.1016/j.chembiol.2018.03.007
- Primary Citation of Related Structures:
5XGV - PubMed Abstract:
Here, we provide structural insights into PyrE3, a flavin-dependent [4 + 2] cyclase that catalyzes trans-decalin formation in the biosynthesis of pyrroindomycins. PyrE3 shares an architecture/domain organization head-to-tail similarity with the members of the family of para-hydroxybenzoate hydroxylase (pHBH)-fold monooxygenases, and possesses a flavin adenine dinucleotide (FAD)-binding domain, a middle domain, and a C-terminal thioredoxin-like domain. The FAD-binding domain forms a central hub of the protein structure, and binds with FAD in a "closed" conformation of pHBH-fold family monooxygenases known for their highly dynamic catalytic processes. FAD plays an essential structural role in PyrE3, where it is amenable to redox change; however, redox change has little effect on [4 + 2] cyclization activity. PyrE3 appears to selectively accommodate a tetramate-containing, linear polyene intermediate in a highly positively charged pocket, which is located at the interface between the FAD-binding domain and the middle domain, and can accelerate trans-decalin formation likely through an endo-selective [4 + 2] transition state.
Organizational Affiliation:
State Key Laboratory of Bioorganic and Natural Products Chemistry, Center for Excellence in Molecular Sciences, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, 345 Lingling Road, Shanghai 200032, China.