Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein
Guo, M., Wang, S., Zhu, Y., Wang, S., Xiong, Z., Yang, J., Xu, Z., Huang, Z.(2017) Nature 546: 436-439
- PubMed: 28448066
- DOI: https://doi.org/10.1038/nature22377
- Primary Citation of Related Structures:
5XBL - PubMed Abstract:
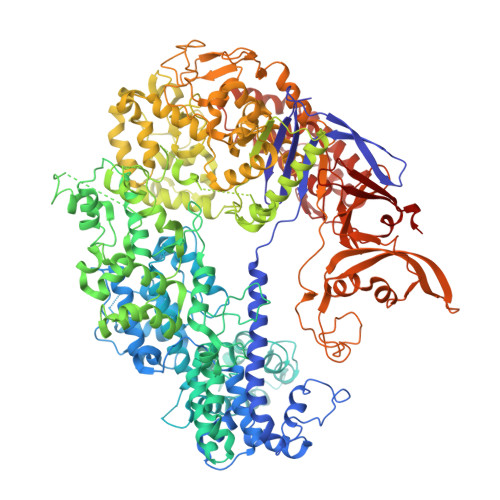
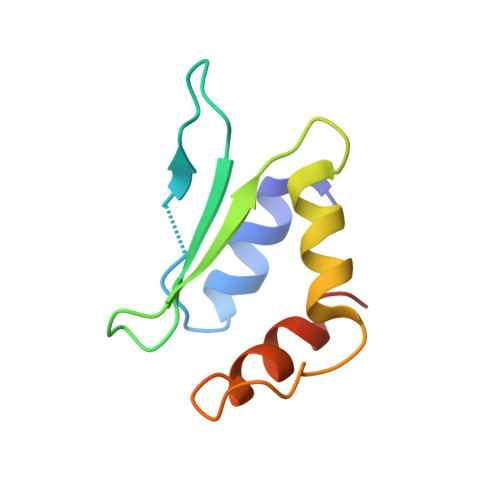
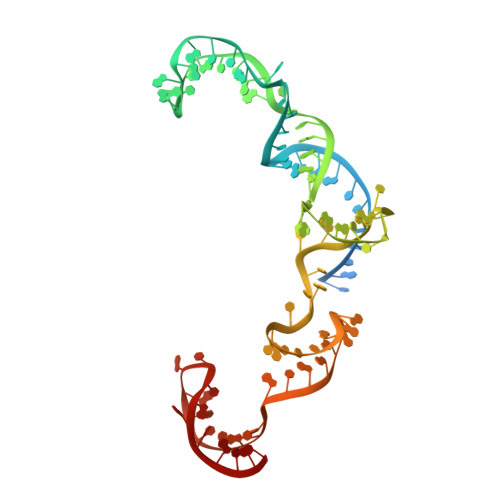
CRISPR-Cas9 systems are bacterial adaptive immune systems that defend against infection by phages. Through the RNA-guided endonuclease activity of Cas9 they degrade double-stranded DNA with a protospacer adjacent motif (PAM) and sequences complementary to the guide RNA. Recently, two anti-CRISPR proteins (AcrIIA2 and AcrIIA4 from Listeria monocytogenes prophages) were identified, both of which inhibit Streptococcus pyogenes Cas9 (SpyCas9) and L. monocytogenes Cas9 activity in bacteria and human cells. However, the mechanism of AcrIIA2- or AcrIIA4-mediated Cas9 inhibition remains unknown. Here we report a crystal structure of SpyCas9 in complex with a single-guide RNA (sgRNA) and AcrIIA4. Our data show that AcrIIA2 and AcrIIA4 interact with SpyCas9 in a sgRNA-dependent manner. The structure reveals that AcrIIA4 inhibits SpyCas9 activity by structurally mimicking the PAM to occupy the PAM-interacting site in the PAM-interacting domain, thereby blocking recognition of double-stranded DNA substrates by SpyCas9. AcrIIA4 further inhibits the endonuclease activity of SpyCas9 by shielding its RuvC active site. Structural comparison reveals that formation of the AcrIIA4-binding site of SpyCas9 is induced by sgRNA binding. Our study reveals the mechanism of SpyCas9 inhibition by AcrIIA4, providing a structural basis for developing 'off-switch' tools for SpyCas9 to avoid unwanted genome edits within cells and tissues.
Organizational Affiliation:
HIT Center for Life Sciences, School of Life Science and Technology, Harbin Institute of Technology, Harbin 150080, China.