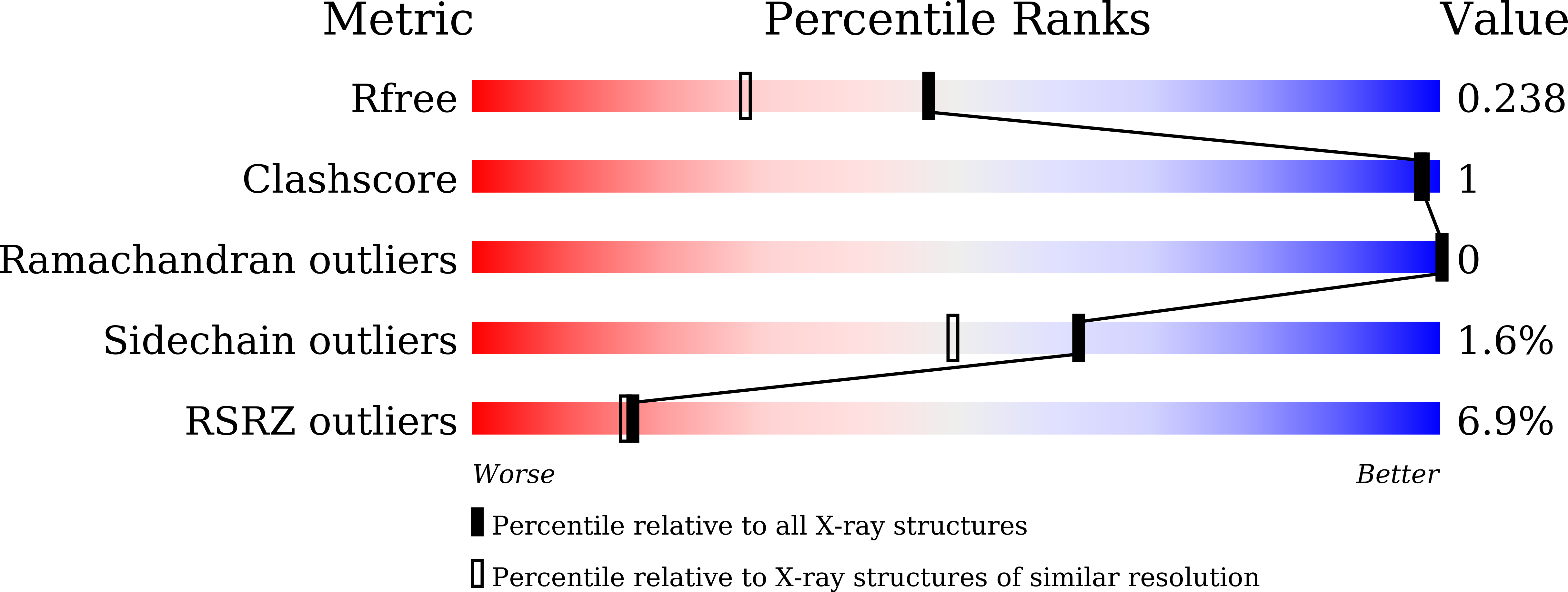
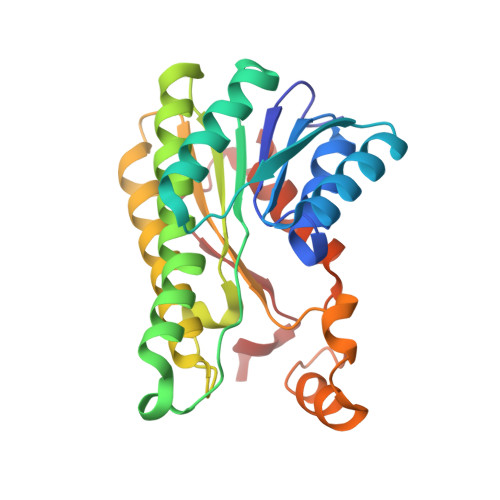
Crystal structure and iterative saturation mutagenesis of ChKRED20 for expanded catalytic scope
Zhao, F.J., Jin, Y., Liu, Z., Guo, C., Li, T.B., Li, Z.Y., Wang, G., Wu, Z.L.(2017) Appl Microbiol Biotechnol 101: 8395-8404
- PubMed: 29067484
- DOI: https://doi.org/10.1007/s00253-017-8556-2
- Primary Citation of Related Structures:
5X8H - PubMed Abstract:
ChKRED20 is an efficient and robust anti-Prelog ketoreductase that can catalyze the reduction of ketones to chiral alcohols as pharmaceutical intermediates with great industrial potential. To overcome its limitation on the bioreduction of ortho-substituted acetophenone derivatives, the X-ray crystal structure of the apo-enzyme of ChKRED20 was determined at a resolution of 1.85 Å and applied to the molecular modeling and reshaping of the catalytic cavity via three rounds of iterative saturation mutagenesis together with alanine scanning and recombination. The mutant Mut3B was achieved with expanded catalytic scope that covered all the nine substrates tested as compared with two substrates for the wild type. It exhibited 13-20-fold elevated k cat /K m values relative to the wild type or to the first gain-of-activity mutant, while retaining excellent stereoselectivity toward seven of the substrates (98-> 99% ee). Another mutant 29G10 displayed complementary selectivity for eight of the ortho-substituted acetophenone derivatives, with six of them delivering excellent stereoselectivity (90-99% ee). Its k cat /K m value toward 1-(2-fluorophenyl)ethanone was 5.6-fold of the wild type. The application of Mut3B in elevated substrate concentrations of 50-100 g/l was demonstrated in 50-ml reactions, achieving 75-> 99% conversion and > 99% ee.
Organizational Affiliation:
Key Laboratory of Environmental and Applied Microbiology, Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, 610041, China.