Ligand binding induced folding of a novel CBM69 starch binding domain
Li, X., Yu, J., Tu, X., Sun, H., Peng, H., Zhang, X.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Amylase | 157 | uncultured bacterium | Mutation(s): 0 Gene Names: Amy-1E | 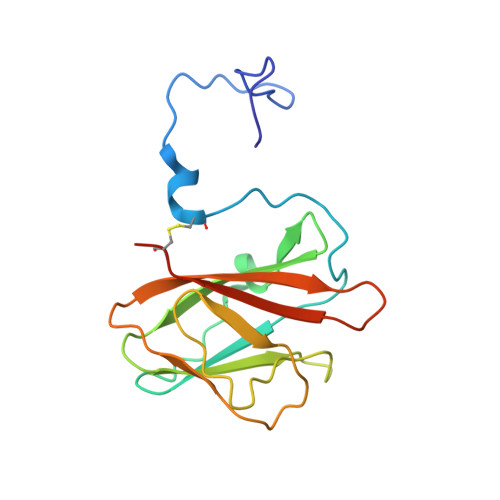 | |
UniProt | |||||
Find proteins for D9MZ14 (uncultured bacterium) Explore D9MZ14 Go to UniProtKB: D9MZ14 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D9MZ14 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Chinese National Natural Science Foundation | China | 31470775 |