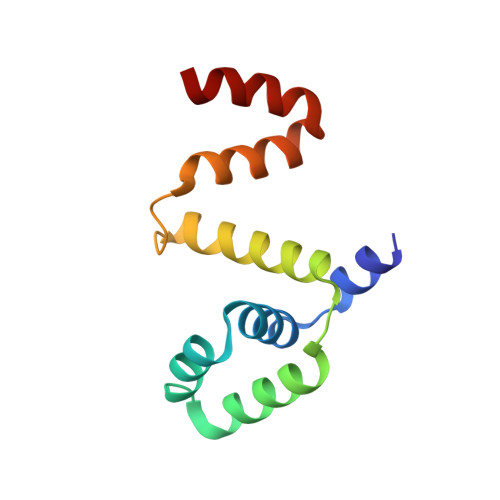
Crystal structure of the FliF-FliG complex from Helicobacter pylori yields insight into the assembly of the motor MS-C ring in the bacterial flagellum
Xue, C., Lam, K.H., Zhang, H., Sun, K., Lee, S.H., Chen, X., Au, S.W.N.(2018) J Biol Chem 293: 2066-2078
- PubMed: 29229777
- DOI: https://doi.org/10.1074/jbc.M117.797936
- Primary Citation of Related Structures:
5WUJ - PubMed Abstract:
The bacterial flagellar motor is a self-assembling supramolecular nanodevice. Its spontaneous biosynthesis is initiated by the insertion of the MS ring protein FliF into the inner membrane, followed by attachment of the switch protein FliG. Assembly of this multiprotein complex is tightly regulated to avoid nonspecific aggregation, but the molecular mechanisms governing flagellar assembly are unclear. Here, we present the crystal structure of the cytoplasmic domain of FliF complexed with the N-terminal domain of FliG (FliF C -FliG N ) from the bacterium Helicobacter pylori Within this complex, FliF C interacted with FliG N through extensive hydrophobic contacts similar to those observed in the FliF C -FliG N structure from the thermophile Thermotoga maritima , indicating conservation of the FliF C -FliG N interaction across bacterial species. Analysis of the crystal lattice revealed that the heterodimeric complex packs as a linear superhelix via stacking of the armadillo repeat-like motifs (ARM) of FliG N Notably, this linear helix was similar to that observed for the assembly of the FliG middle domain. We validated the in vivo relevance of the FliG N stacking by complementation studies in Escherichia coli Furthermore, structural comparison with apo FliG from the thermophile Aquifex aeolicus indicated that FliF regulates the conformational transition of FliG and exposes the complementary ARM-like motifs of FliG N , containing conserved hydrophobic residues. FliF apparently both provides a template for FliG polymerization and spatiotemporally controls subunit interactions within FliG. Our findings reveal that a small protein fold can serve as a versatile building block to assemble into a multiprotein machinery of distinct shapes for specific functions.
Organizational Affiliation:
From the Center for Protein Science and Crystallography, School of Life Sciences, The Chinese University of Hong Kong, Shatin, Hong Kong, China.