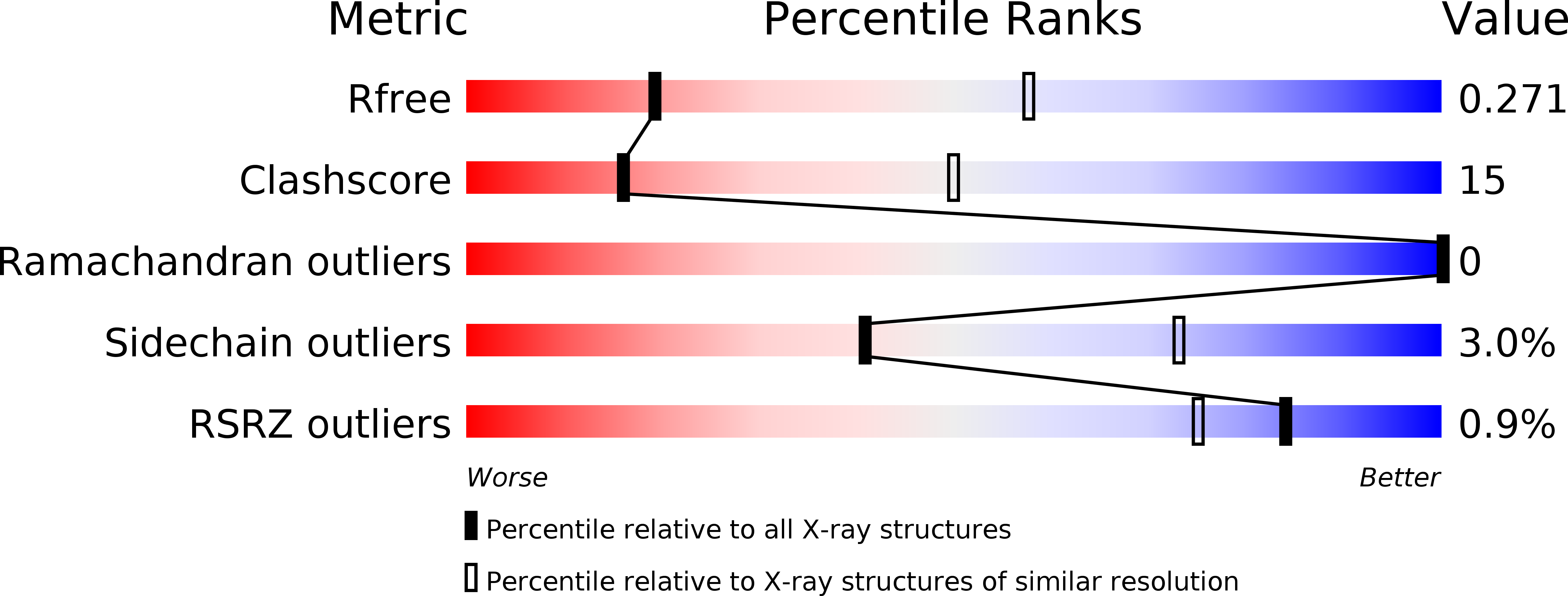
Structural and Biochemical Characterization of Apicomplexan Inorganic Pyrophosphatases
Jamwal, A., Yogavel, M., Abdin, M.Z., Jain, S.K., Sharma, A.(2017) Sci Rep 7: 5255-5255
- PubMed: 28701714
- DOI: https://doi.org/10.1038/s41598-017-05234-y
- Primary Citation of Related Structures:
5WRT, 5WRU - PubMed Abstract:
Inorganic pyrophosphatases (PPase) participate in energy cycling and they are essential for growth and survival of organisms. Here we report extensive structural and functional characterization of soluble PPases from the human parasites Plasmodium falciparum (PfPPase) and Toxoplasma gondii (TgPPase). Our results show that PfPPase is a cytosolic enzyme whose gene expression is upregulated during parasite asexual stages. Cambialistic PfPPase actively hydrolyzes linear short chain polyphosphates like PP i , polyP 3 and ATP in the presence of Zn 2+ . A remarkable new feature of PfPPase is the low complexity asparagine-rich N-terminal region that mediates its dimerization. Deletion of N-region has an unexpected and substantial effect on the stability of PfPPase domain, resulting in aggregation and significant loss of enzyme activity. Significantly, the crystal structures of PfPPase and TgPPase reveal unusual and unprecedented dimeric organizations and provide new fundamental insights into the variety of oligomeric assemblies possible in eukaryotic inorganic PPases.
Organizational Affiliation:
Molecular Medicine Group, International Centre for Genetic Engineering and Biotechnology, Aruna Asaf Ali Marg, New Delhi, 110067, India.