The virulence-associated protein HsvA from the fire blight pathogen Erwinia amylovora is a polyamine amidinotransferase.
Shanker, S., Schaefer, G.K., Barnhart, B.K., Wallace-Kneale, V.L., Chang, D., Coyle, T.J., Metzler, D.A., Huang, J., Lawton, J.A.(2017) J Biol Chem 292: 21366-21380
- PubMed: 29123034
- DOI: https://doi.org/10.1074/jbc.M117.815951
- Primary Citation of Related Structures:
5WPI - PubMed Abstract:
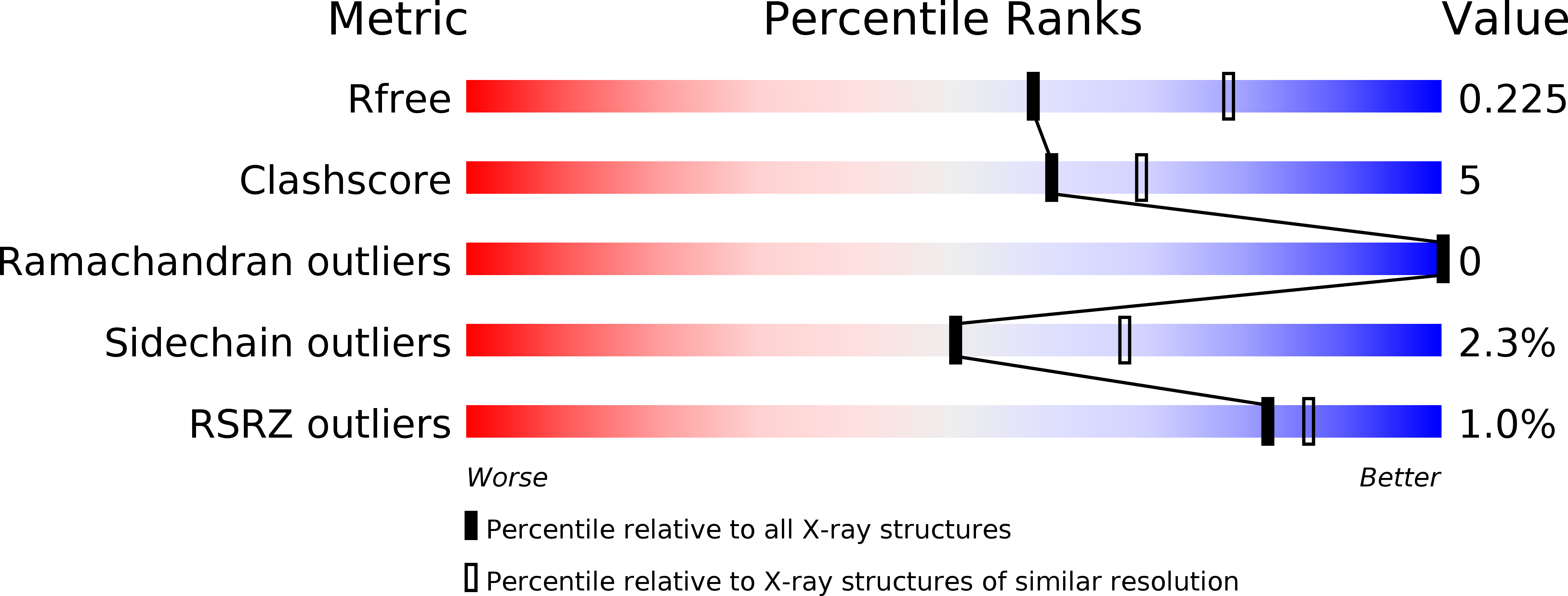
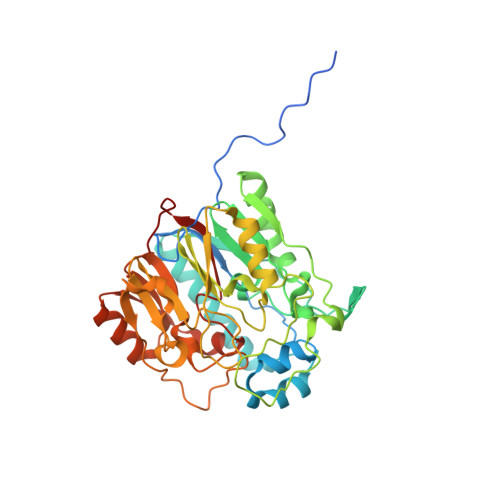
Studies of virulence determinants in the bacterial phytopathogen Erwinia amylovora , the cause of devastating fire blight disease in apple and pear, have shown that HsvA, a putative amidinotransferase enzyme located in the Hrp pathogenicity island, is required for systemic infection in apple. However, the mechanism by which HsvA contributes to virulence is unclear. To investigate the role of HsvA in virulence, we carried out a series of biochemical and structural studies to characterize the amidinotransferase activity of HsvA. We found that HsvA displays a preference for linear aliphatic polyamines as the amidino acceptor substrate, especially for spermidine and putrescine ( K m values of 33 μm and 3.9 mm, respectively). The three-dimensional structure, determined at 2.30 Å resolution using X-ray crystallography, revealed that the overall architecture of HsvA is similar to that of the human arginine-glycine amidinotransferase in the creatine biosynthesis pathway. The active site is located in the core of the protein at the base of a long, narrow substrate access channel. Specific amino acids near the entrance of the channel may serve as major determinants of the substrate specificity, including a glutamate residue at the rim of the channel entrance that appears to be positioned to interact with the distal primary amine in the putrescine substrate as well as the internal and distal amines in the spermidine substrate. These results suggest potential in vivo functions for HsvA as a virulence factor in fire blight and may also provide a basis for strategies to control fire blight by inhibiting HsvA activity.
Organizational Affiliation:
From the Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, Texas 77030 and.