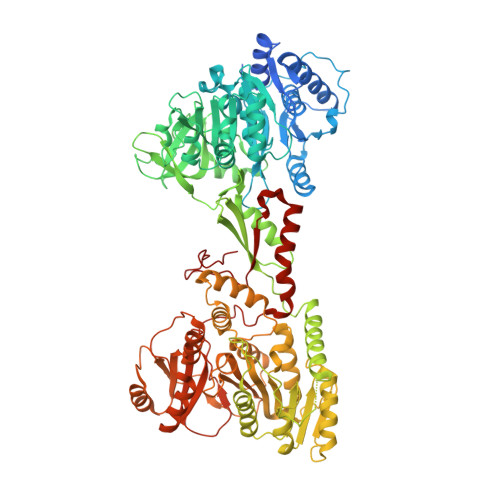
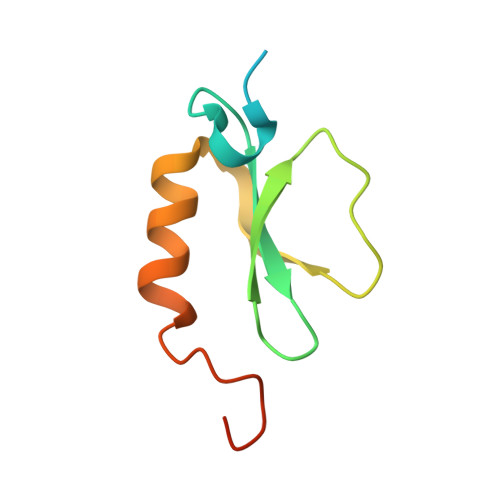
Structural basis for backbone N-methylation by an interrupted adenylation domain.
Mori, S., Pang, A.H., Lundy, T.A., Garzan, A., Tsodikov, O.V., Garneau-Tsodikova, S.(2018) Nat Chem Biol 14: 428-430
- PubMed: 29556104
- DOI: https://doi.org/10.1038/s41589-018-0014-7
- Primary Citation of Related Structures:
5WMM - PubMed Abstract:
Interrupted adenylation domains are enigmatic fusions, in which one enzyme is inserted into another to form a highly unusual bifunctional enzyme. We present the first crystal structure of an interrupted adenylation domain that reveals a unique embedded methyltransferase. The structure and functional data provide insight into how these enzymes N-methylate amino acid precursors en route to nonribosomal peptides.
Organizational Affiliation:
Department of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky, Lexington, KY, USA.