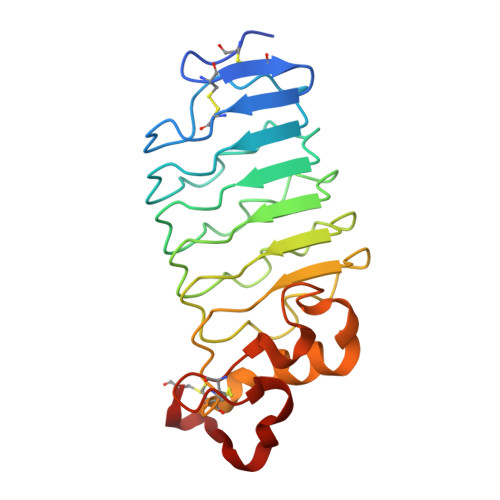
Crystal structure of an anti-idiotype variable lymphocyte receptor.
Collins, B.C., Nakahara, H., Acharya, S., Cooper, M.D., Herrin, B.R., Wilson, I.A.(2017) Acta Crystallogr F Struct Biol Commun 73: 682-687
- PubMed: 29199989
- DOI: https://doi.org/10.1107/S2053230X1701620X
- Primary Citation of Related Structures:
5WK4 - PubMed Abstract:
Variable lymphocyte receptors (VLRs), the leucine-rich repeat (LRR)-based antigen receptors of jawless fish, have great utility in a wide variety of biochemical and biological applications, similar to classical Ig-based antibodies. VLR-based reagents may be particularly useful when traditional antibodies are not available. An anti-idiotype lamprey VLR, VLR39, has previously been identified that recognizes the heavy-chain CDR3 of the B-cell receptor (BCR) of a leukemic clone from a patient with chronic lymphocytic leukemia (CLL). VLR39 was used successfully to track the re-emergence of this clone in the patient following chemotherapy. Here, the crystal structure of VLR39 is presented at 1.5 Å resolution and compared with those of other protein-specific VLRs. VLR39 adopts a curved solenoid fold and exhibits substantial structural similarity to other protein-binding VLRs. VLR39 has a short LRRCT loop that protrudes outwards away from the concave face and is similar to those of its protein-specific VLR counterparts. Analysis of the VLR39-BCR interaction by size-exclusion chromatography and biolayer interferometry using the scFv version of the BCR confirms that VLR39 recognizes the BCR Fv region. Such VLR-based reagents may be useful for identifying and monitoring leukemia in CLL patients and in other clinical diagnostic assays.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology and the Skaggs Institute for Chemical Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.