Structure of the Marine Siphovirus TW1: Evolution of Capsid-Stabilizing Proteins and Tail Spikes.
Wang, Z., Hardies, S.C., Fokine, A., Klose, T., Jiang, W., Cho, B.C., Rossmann, M.G.(2018) Structure 26: 238-248.e3
- PubMed: 29290487
- DOI: https://doi.org/10.1016/j.str.2017.12.001
- Primary Citation of Related Structures:
5WK1 - PubMed Abstract:
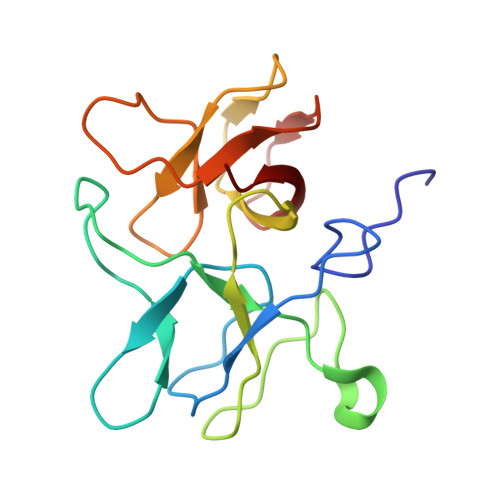
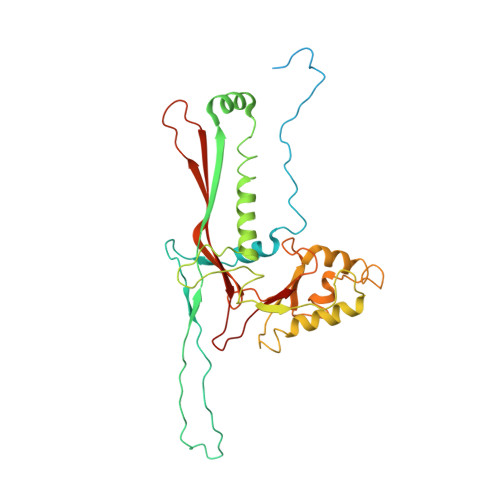
Marine bacteriophage TW1 belongs to the Siphoviridae family and infects Pseudoalteromonas phenolica. Mass spectrometry analysis has identified 16 different proteins in the TW1 virion. Functions of most of these proteins have been predicted by bioinformatic methods. A 3.6 Å resolution cryoelectron microscopy map of the icosahedrally averaged TW1 head showed the atomic structures of the major capsid protein, gp57 ∗ , and the capsid-stabilizing protein, gp56. The gp57 ∗ structure is similar to that of the phage HK97 capsid protein. The gp56 protein has two domains, each having folds similar to that of the N-terminal part of phage λ gpD, indicating a common ancestry. The first gp56 domain clamps adjacent capsomers together, whereas the second domain is required for trimerization. A 6-fold-averaged reconstruction of the distal part of the tail showed that TW1 has six tail spikes, which are unusual for siphophages but are similar to the podophages P22 and Sf6, suggesting a common evolutionary origin of these spikes.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, IN 47907, USA.