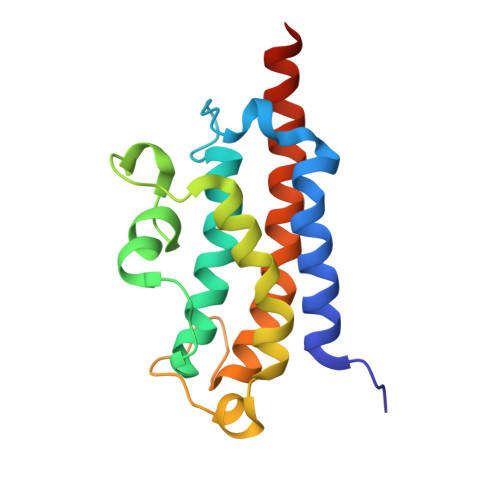
Structure and function of the bacillithiol-S-transferase BstA from Staphylococcus aureus.
Francis, J.W., Royer, C.J., Cook, P.D.(2018) Protein Sci 27: 898-902
- PubMed: 29417696
- DOI: https://doi.org/10.1002/pro.3384
- Primary Citation of Related Structures:
5WK0 - PubMed Abstract:
Bacillithiol is a low-molecular weight thiol produced by many gram-positive organisms, including Staphylococcus aureus and Bacillus anthracis. It is the major thiol responsible for maintaining redox homeostasis and cellular detoxification, including inactivation of the antibiotic fosfomycin. The metal-dependent bacillithiol transferase BstA is likely involved in these sorts of detoxification processes, but the exact substrates and enzyme mechanism have not been identified. Here we report the 1.34 Å resolution X-ray crystallographic structure of BstA from S. aureus. Our structure confirms that BstA belongs to the YfiT-like metal-dependent hydrolase superfamily. Like YfiT, our structure contains nickel within its active site, but our functional data suggest that BstA utilizes zinc for activity. Although BstA and YfiT both contain a core four helix bundle and coordinate their metal ions in the same fashion, significant differences between the protein structures are described here.
Organizational Affiliation:
Department of Chemistry, Grand Valley State University, Allendale, Michigan, 49401.