Mis16 Switches Function from a Histone H4 Chaperone to a CENP-ACnp1-Specific Assembly Factor through Eic1 Interaction.
An, S., Koldewey, P., Chik, J., Subramanian, L., Cho, U.S.(2018) Structure 26: 960
- PubMed: 29804820
- DOI: https://doi.org/10.1016/j.str.2018.04.012
- Primary Citation of Related Structures:
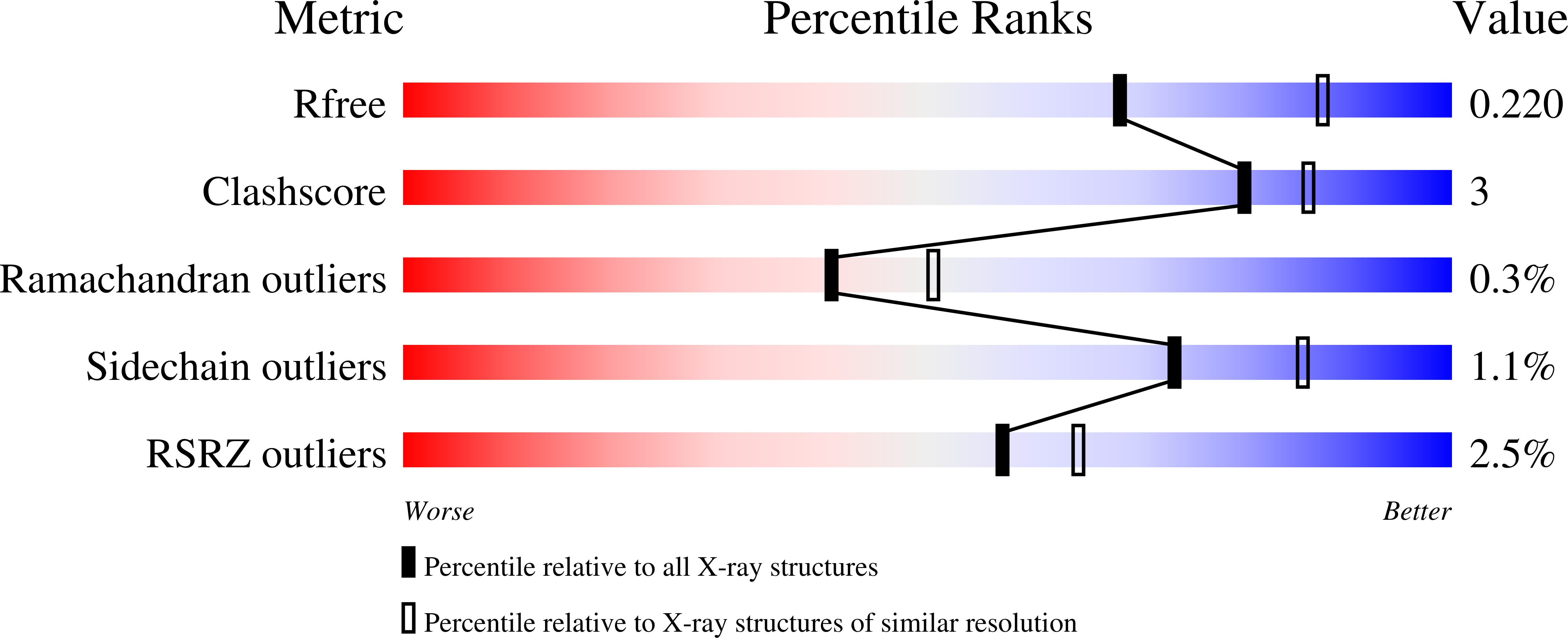
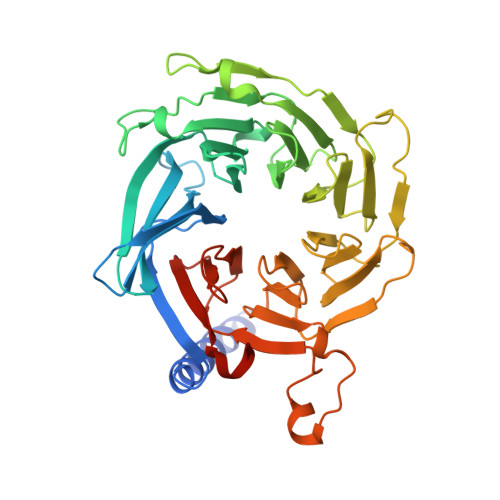
5WJC - PubMed Abstract:
The Mis18 complex, composed of Mis16, Eic1, and Mis18 in fission yeast, selectively deposits the centromere-specific histone H3 variant, CENP-A Cnp1 , at centromeres. How the intact Mis18 holo-complex oligomerizes and how Mis16, a well-known ubiquitous histone H4 chaperone, plays a centromere-specific role in the Mis18 holo-complex, remain unclear. Here, we report the stoichiometry of the intact Mis18 holo-complex as (Mis16) 2 :(Eic1) 2 :(Mis18) 4 using analytical ultracentrifugation. We further determine the crystal structure of Schizosaccharomyces pombe Mis16 in complex with the C-terminal portion of Eic1 (Eic1-CT). Notably, Mis16 accommodates Eic1-CT through the binding pocket normally occupied by histone H4, indicating that Eic1 and H4 compete for the same binding site, providing a mechanism for Mis16 to switch its binding partner from histone H4 to Eic1. Thus, our analyses not only determine the stoichiometry of the intact Mis18 holo-complex but also uncover the molecular mechanism by which Mis16 plays a centromere-specific role through Eic1 association.
Organizational Affiliation:
Department of Biological Chemistry, University of Michigan Medical School, 1150 W. Medical Center Drive, SPC 5606, Ann Arbor, MI 48109, USA.