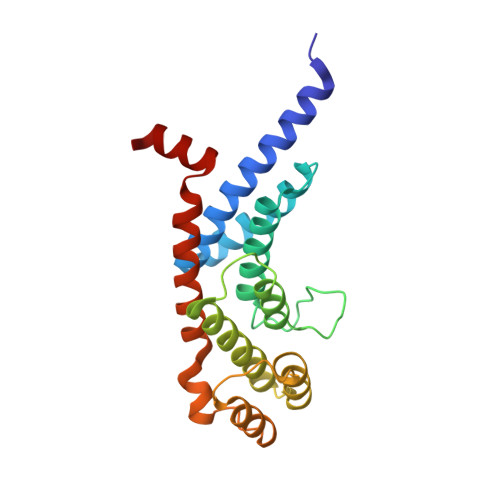
Dissecting the telomere-inner nuclear membrane interface formed in meiosis.
Pendlebury, D.F., Fujiwara, Y., Tesmer, V.M., Smith, E.M., Shibuya, H., Watanabe, Y., Nandakumar, J.(2017) Nat Struct Mol Biol 24: 1064-1072
- PubMed: 29083414
- DOI: https://doi.org/10.1038/nsmb.3493
- Primary Citation of Related Structures:
5WIR - PubMed Abstract:
Tethering telomeres to the inner nuclear membrane (INM) allows homologous chromosome pairing during meiosis. The meiosis-specific protein TERB1 binds the telomeric protein TRF1 to establish telomere-INM connectivity and is essential for mouse fertility. Here we solve the structure of the human TRF1-TERB1 interface to reveal the structural basis for telomere-INM linkage. Disruption of this interface abrogates binding and compromises telomere-INM attachment in mice. An embedded CDK-phosphorylation site within the TRF1-binding region of TERB1 provides a mechanism for cap exchange, a late-pachytene phenomenon involving the dissociation of the TRF1-TERB1 complex. Indeed, further strengthening this interaction interferes with cap exchange. Finally, our biochemical analysis implicates distinct complexes for telomere-INM tethering and chromosome-end protection during meiosis. Our studies unravel the structure, stoichiometry, and physiological implications underlying telomere-INM tethering, thereby providing unprecedented insights into the unique function of telomeres in meiosis.
Organizational Affiliation:
Department of Molecular, Cellular, and Developmental Biology, University of Michigan, Ann Arbor, Michigan, USA.