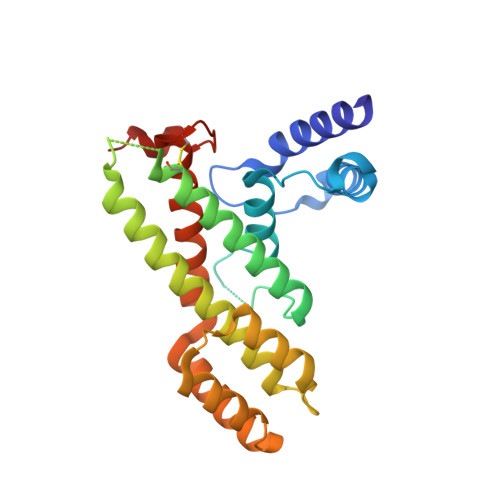
Structural Basis for Shelterin Bridge Assembly.
Kim, J.K., Liu, J., Hu, X., Yu, C., Roskamp, K., Sankaran, B., Huang, L., Komives, E.A., Qiao, F.(2017) Mol Cell 68: 698-714.e5
- PubMed: 29149597
- DOI: https://doi.org/10.1016/j.molcel.2017.10.032
- Primary Citation of Related Structures:
5WE0, 5WE1, 5WE2 - PubMed Abstract:
Telomere elongation through telomerase enables chromosome survival during cellular proliferation. The conserved multifunctional shelterin complex associates with telomeres to coordinate multiple telomere activities, including telomere elongation by telomerase. Similar to the human shelterin, fission yeast shelterin is composed of telomeric sequence-specific double- and single-stranded DNA-binding proteins, Taz1 and Pot1, respectively, bridged by Rap1, Poz1, and Tpz1. Here, we report the crystal structure of the fission yeast Tpz1 475-508 -Poz1-Rap1 467-496 complex that provides the structural basis for shelterin bridge assembly. Biochemical analyses reveal that shelterin bridge assembly is a hierarchical process in which Tpz1 binding to Poz1 elicits structural changes in Poz1, allosterically promoting Rap1 binding to Poz1. Perturbation of the cooperative Tpz1-Poz1-Rap1 assembly through mutation of the "conformational trigger" in Poz1 leads to unregulated telomere lengthening. Furthermore, we find that the human shelterin counterparts TPP1-TIN2-TRF2 also assemble hierarchically, indicating cooperativity as a conserved driving force for shelterin assembly.
Organizational Affiliation:
Department of Biological Chemistry, School of Medicine, University of California, Irvine, Irvine, CA 92697-1700, USA.