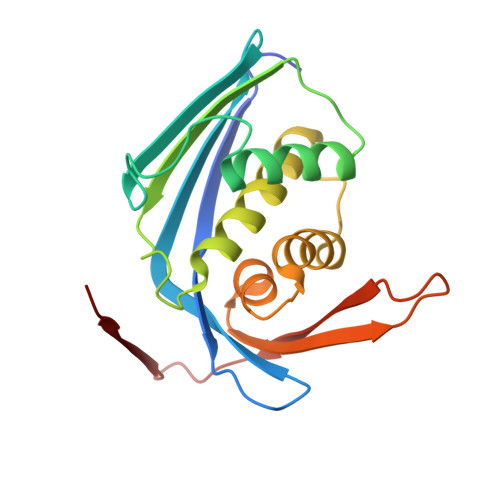
Reduction of Feedback Inhibition in Homoserine Kinase (ThrB) ofCorynebacterium glutamicumEnhances l-Threonine Biosynthesis.
Petit, C., Kim, Y., Lee, S.K., Brown, J., Larsen, E., Ronning, D.R., Suh, J.W., Kang, C.M.(2018) ACS Omega 3: 1178-1186
- PubMed: 30023797
- DOI: https://doi.org/10.1021/acsomega.7b01597
- Primary Citation of Related Structures:
5WAS, 5WAT - PubMed Abstract:
l-Threonine is an important supplement in the food industry. It is currently produced through fermentation of Escherichia coli but requires additional purification steps to remove E. coli endotoxin. To avoid these steps, it is desirable to use Corynebacterium glutamicum , a microorganism generally regarded as safe. Engineering of C. glutamicum to increase production of l-threonine has mainly focused on gene regulation as well as l-threonine export or carbon flux depletion. In this study, we focus on the negative feedback inhibition produced by l-threonine on the enzyme homoserine kinase (ThrB). Although l-threonine binds to allosteric sites of aspartate kinase (LysC) and homoserine dehydrogenase (Hom), serving as a noncompetitive inhibitor, it acts as a competitive inhibitor on ThrB. This is problematic when attempting to engineer enzymes that are nonresponsive to increasing cellular concentrations of l-threonine. Using primary structure alignment as well as analysis of the Methanocaldococcus jannaschii ThrB ( Mja ThrB) active site in complex with l-threonine (inhibitor of ThrB) and l-homoserine (substrate of ThrB), a conserved active-site alanine residue (A20) in C. glutamicum ThrB ( Cgl ThrB) was predicted to be important for differential interactions with l-threonine and l-homoserine. Through site-directed mutagenesis, we show that one variant of C. glutamicum ThrB, Cgl ThrB-A20G, retains wild-type enzymatic activity, with dramatically decreased feedback inhibition by l-threonine. Additionally, by solving the first Corynebacterium X-ray crystal structure of homoserine kinase, we can confirm that the changes in l-threonine affinity to the Cgl ThrB-A20G active site derive from loss of van der Waals interactions.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Toledo, 2801 W. Bancroft Street, Toledo, Ohio 43606, United States.