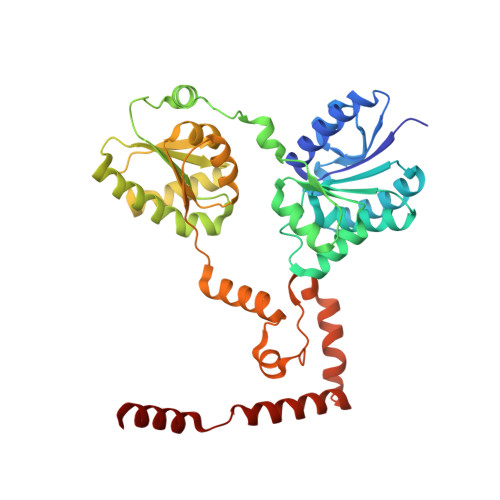
Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli.
Bohl, T.E., Shi, K., Lee, J.K., Aihara, H.(2018) Nat Commun 9: 377-377
- PubMed: 29371662
- DOI: https://doi.org/10.1038/s41467-017-02712-9
- Primary Citation of Related Structures:
5W8N, 5W8S, 5W8X - PubMed Abstract:
Most Gram-negative bacteria are surrounded by a glycolipid called lipopolysaccharide (LPS), which forms a barrier to hydrophobic toxins and, in pathogenic bacteria, is a virulence factor. During LPS biosynthesis, a membrane-associated glycosyltransferase (LpxB) forms a tetra-acylated disaccharide that is further acylated to form the membrane anchor moiety of LPS. Here we solve the structure of a soluble and catalytically competent LpxB by X-ray crystallography. The structure reveals that LpxB has a glycosyltransferase-B family fold but with a highly intertwined, C-terminally swapped dimer comprising four domains. We identify key catalytic residues with a product, UDP, bound in the active site, as well as clusters of hydrophobic residues that likely mediate productive membrane association or capture of lipidic substrates. These studies provide the basis for rational design of antibiotics targeting a crucial step in LPS biosynthesis.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota Twin Cities, Minneapolis, MN, 55455, USA.