Biochemical and Structural Analyses of Two Cryptic Esterases in Bacteroides intestinalis and their Synergistic Activities with Cognate Xylanases.
Wefers, D., Cavalcante, J.J.V., Schendel, R.R., Deveryshetty, J., Wang, K., Wawrzak, Z., Mackie, R.I., Koropatkin, N.M., Cann, I.(2017) J Mol Biol 429: 2509-2527
- PubMed: 28669823
- DOI: https://doi.org/10.1016/j.jmb.2017.06.017
- Primary Citation of Related Structures:
5VOL - PubMed Abstract:
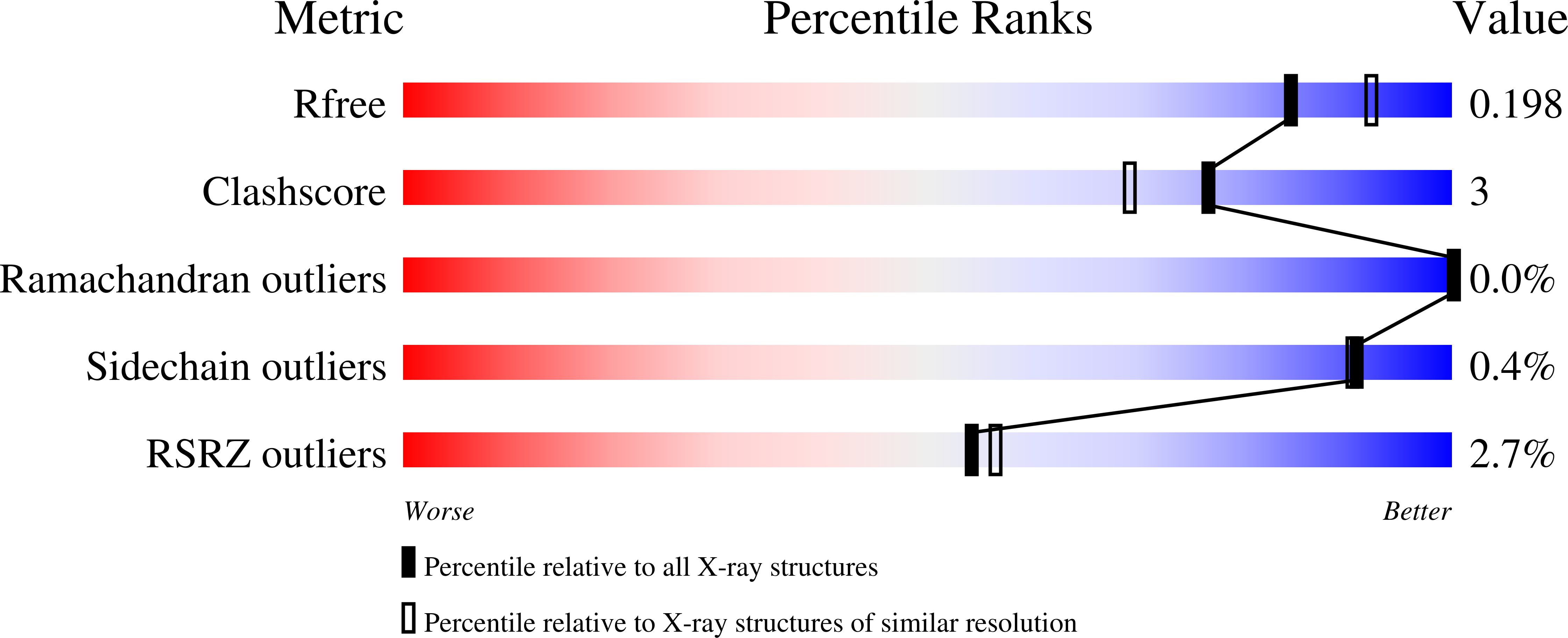
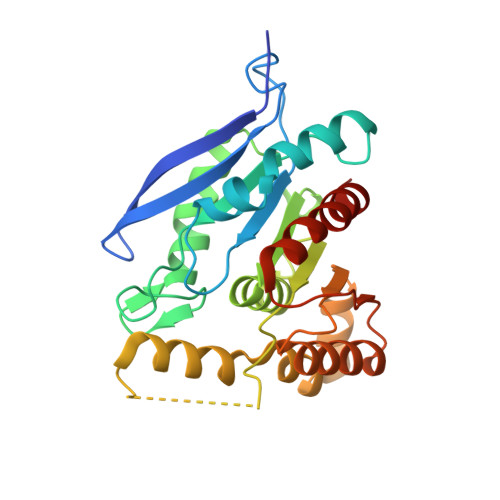
Arabinoxylans are constituents of the human diet. Although not utilizable by the human host, they can be fermented by colonic bacteria. The arabinoxylan backbone is decorated with arabinose side chains that may be substituted with ferulic acid, thus limiting depolymerization to fermentable sugars. We investigated the polypeptides encoded by two genes upregulated during growth of the colonic bacterium Bacteroides intestinalis on wheat arabinoxylan. The recombinant proteins, designated BiFae1A and BiFae1B, were functionally assigned esterase activities. Both enzymes were active on acetylated substrates, although each showed a higher ferulic acid esterase activity on methyl-ferulate. BiFae1A showed a catalytic efficiency of 12mM s -1 on para-nitrophenyl-acetate, and on methyl-ferulate, the value was 27 times higher. BiFae1B showed low catalytic efficiencies for both substrates. Furthermore, the two enzymes released ferulic acid from various structural elements, and NMR spectroscopy indicated complete de-esterification of arabinoxylan oligosaccharides from wheat bran. BiFae1A is a tetramer based on the crystal structure, whereas BiFae1B is a dimer in solution based on size exclusion chromatography. The structure of BiFae1A was solved to 1.98Å resolution, and two tetramers were observed in the asymmetric unit. A flexible loop that may act as a hinge over the active site and likely coordinates critical interactions with the substrate was prominent in BiFae1A. Sequence alignments of the esterase domains in BiFae1B with the feruloyl esterase from Clostridium thermocellum suggest that both domains lack the flexible hinge in BiFae1A, an observation that may partly provide a molecular basis for the differences in activities in the two esterases.
Organizational Affiliation:
Carl R. Woese Institute for Genomic Biology (Microbiome Metabolic Engineering Theme), University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA; Department of Animal Science, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.