A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription.
Sdano, M.A., Fulcher, J.M., Palani, S., Chandrasekharan, M.B., Parnell, T.J., Whitby, F.G., Formosa, T., Hill, C.P.(2017) Elife 6
- PubMed: 28826505
- DOI: https://doi.org/10.7554/eLife.28723
- Primary Citation of Related Structures:
5VKL, 5VKO - PubMed Abstract:
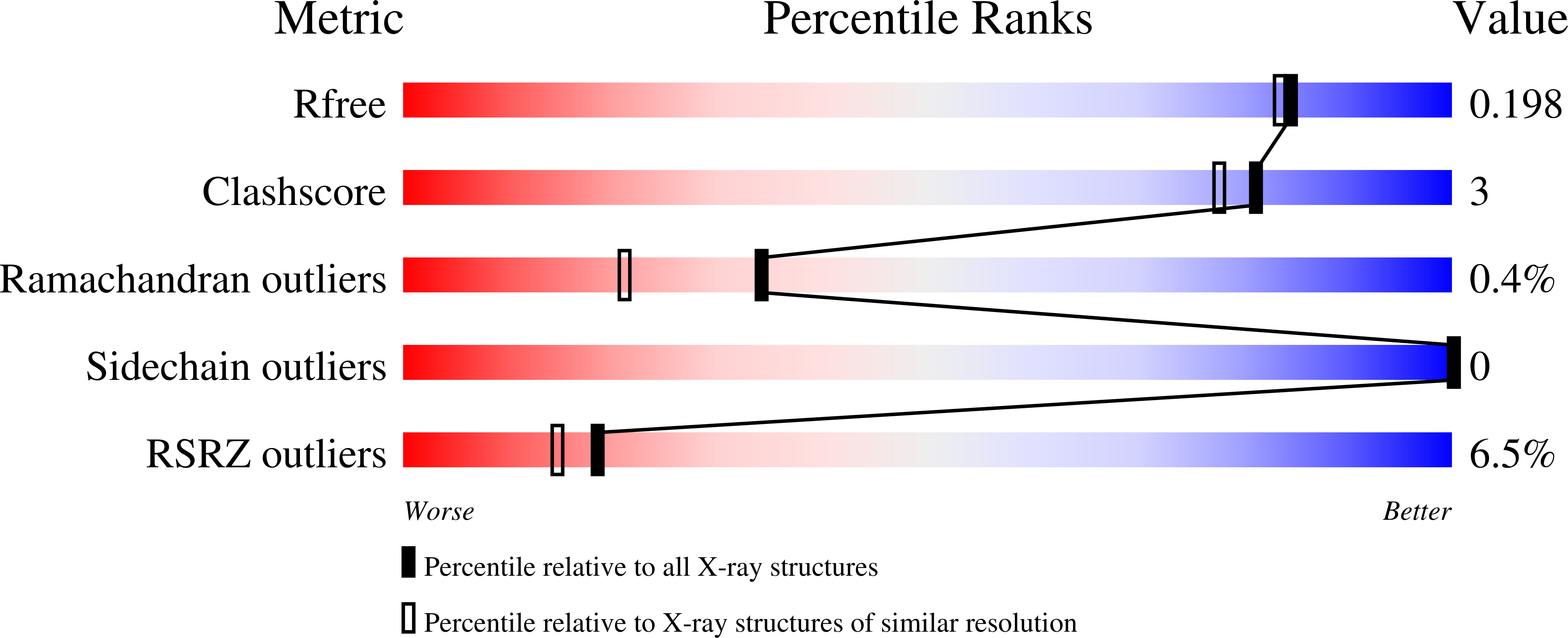
We determined that the tandem SH2 domain of S. cerevisiae Spt6 binds the linker region of the RNA polymerase II subunit Rpb1 rather than the expected sites in its heptad repeat domain. The 4 nM binding affinity requires phosphorylation at Rpb1 S1493 and either T1471 or Y1473. Crystal structures showed that pT1471 binds the canonical SH2 pY site while pS1493 binds an unanticipated pocket 70 Å distant. Remarkably, the pT1471 phosphate occupies the phosphate-binding site of a canonical pY complex, while Y1473 occupies the position of a canonical pY side chain, with the combination of pT and Y mimicking a pY moiety. Biochemical data and modeling indicate that pY1473 can form an equivalent interaction, and we find that pT1471/pS1493 and pY1473/pS1493 combinations occur in vivo. ChIP-seq and genetic analyses demonstrate the importance of these interactions for recruitment of Spt6 to sites of transcription and for the maintenance of repressive chromatin.
Organizational Affiliation:
Department of Biochemistry, University of Utah School of Medicine, Salt Lake City, United States.