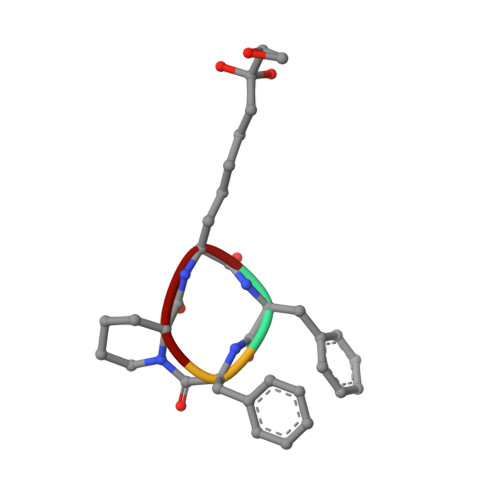
Binding of the Microbial Cyclic Tetrapeptide Trapoxin A to the Class I Histone Deacetylase HDAC8.
Porter, N.J., Christianson, D.W.(2017) ACS Chem Biol 12: 2281-2286
- PubMed: 28846375
- DOI: https://doi.org/10.1021/acschembio.7b00330
- Primary Citation of Related Structures:
5VI6 - PubMed Abstract:
Trapoxin A is a microbial cyclic tetrapeptide that is an essentially irreversible inhibitor of class I histone deacetylases (HDACs). The inhibitory warhead is the α,β-epoxyketone side-chain of (2S,9S)-2-amino-8-oxo-9,10-epoxydecanoic acid (l-Aoe), which mimics the side-chain of the HDAC substrate acetyl-l-lysine. We now report the crystal structure of the HDAC8-trapoxin A complex at 1.24 Å resolution, revealing that the ketone moiety of l-Aoe undergoes nucleophilic attack to form a zinc-bound tetrahedral gem-diolate that mimics the tetrahedral intermediate and its flanking transition states in catalysis. Mass spectrometry, activity measurements, and isothermal titration calorimetry confirm that trapoxin A binds tightly (K d = 3 ± 1 nM) and does not covalently modify the enzyme, so the epoxide moiety of l-Aoe remains intact. Comparison of the HDAC8-trapoxin A complex with the HDAC6-HC toxin complex provides new insight regarding the inhibitory potency of l-Aoe-containing natural products against class I and class II HDACs.
Organizational Affiliation:
Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania , Philadelphia, Pennsylvania 19104-6323, United States.