Structural and Functional Characterization of a Short-Chain Flavodoxin Associated with a Noncanonical 1,2-Propanediol Utilization Bacterial Microcompartment.
Plegaria, J.S., Sutter, M., Ferlez, B., Aussignargues, C., Niklas, J., Poluektov, O.G., Fromwiller, C., TerAvest, M., Utschig, L.M., Tiede, D.M., Kerfeld, C.A.(2017) Biochemistry 56: 5679-5690
- PubMed: 28956602
- DOI: https://doi.org/10.1021/acs.biochem.7b00682
- Primary Citation of Related Structures:
5VEG - PubMed Abstract:
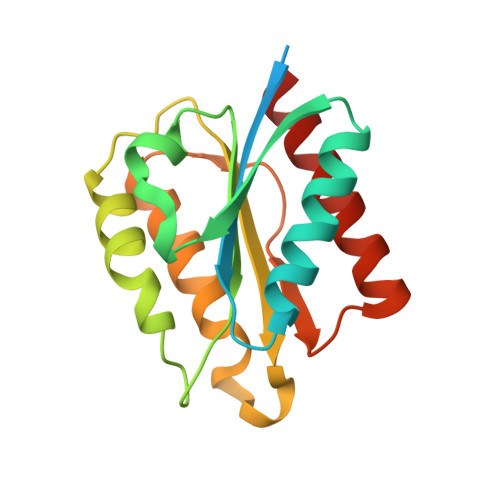
Bacterial microcompartments (BMCs) are proteinaceous organelles that encapsulate enzymes involved in CO 2 fixation (carboxysomes) or carbon catabolism (metabolosomes). Metabolosomes share a common core of enzymes and a distinct signature enzyme for substrate degradation that defines the function of the BMC (e.g., propanediol or ethanolamine utilization BMCs, or glycyl-radical enzyme microcompartments). Loci encoding metabolosomes also typically contain genes for proteins that support organelle function, such as regulation, transport of substrate, and cofactor (e.g., vitamin B 12 ) synthesis and recycling. Flavoproteins are frequently among these ancillary gene products, suggesting that these redox active proteins play an undetermined function in many metabolosomes. Here, we report the first characterization of a BMC-associated flavodoxin (Fld1C), a small flavoprotein, derived from the noncanonical 1,2-propanediol utilization BMC locus (PDU1C) of Lactobacillus reuteri. The 2.0 Å X-ray structure of Fld1C displays the α/β flavodoxin fold, which noncovalently binds a single flavin mononucleotide molecule. Fld1C is a short-chain flavodoxin with redox potentials of -240 ± 3 mV oxidized/semiquinone and -344 ± 1 mV semiquinone/hydroquinone versus the standard hydrogen electrode at pH 7.5. It can participate in an electron transfer reaction with a photoreductant to form a stable semiquinone species. Collectively, our structural and functional results suggest that PDU1C BMCs encapsulate Fld1C to store and transfer electrons for the reactivation and/or recycling of the B 12 cofactor utilized by the signature enzyme.
Organizational Affiliation:
MSU-DOE Plant Research Laboratory, Michigan State University , East Lansing, Michigan 48824, United States.