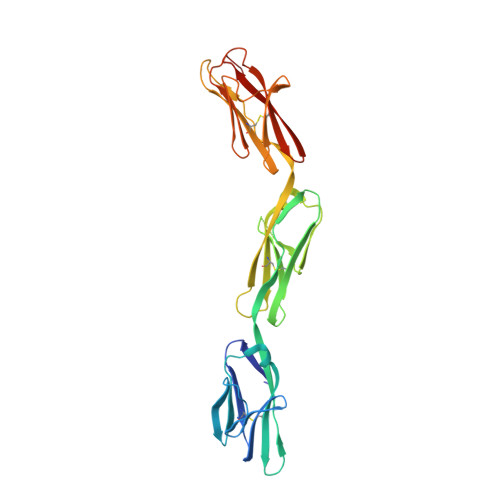
Inactivating mutations and X-ray crystal structure of the tumor suppressor OPCML reveal cancer-associated functions.
Birtley, J.R., Alomary, M., Zanini, E., Antony, J., Maben, Z., Weaver, G.C., Von Arx, C., Mura, M., Marinho, A.T., Lu, H., Morecroft, E.V.N., Karali, E., Chayen, N.E., Tate, E.W., Jurewicz, M., Stern, L.J., Recchi, C., Gabra, H.(2019) Nat Commun 10: 3134-3134
- PubMed: 31316070
- DOI: https://doi.org/10.1038/s41467-019-10966-8
- Primary Citation of Related Structures:
5UV6 - PubMed Abstract:
OPCML, a tumor suppressor gene, is frequently silenced epigenetically in ovarian and other cancers. Here we report, by analysis of databases of tumor sequences, the observation of OPCML somatic missense mutations from various tumor types and the impact of these mutations on OPCML function, by solving the X-ray crystal structure of this glycoprotein to 2.65 Å resolution. OPCML consists of an extended arrangement of three immunoglobulin-like domains and homodimerizes via a network of contacts between membrane-distal domains. We report the generation of a panel of OPCML variants with representative clinical mutations and demonstrate clear phenotypic effects in vitro and in vivo including changes to anchorage-independent growth, interaction with activated cognate receptor tyrosine kinases, cellular migration, invasion in vitro and tumor growth in vivo. Our results suggest that clinically occurring somatic missense mutations in OPCML have the potential to contribute to tumorigenesis in a variety of cancers.
Organizational Affiliation:
Department of Pathology, University of Massachusetts Medical School, Worcester, MA, 01655, USA.